Genome modifications and cloning using a conjugally transferable recombineering system
- PMID: 28352570
- PMCID: PMC4980705
- DOI: 10.1016/j.btre.2015.08.005
Genome modifications and cloning using a conjugally transferable recombineering system
Abstract
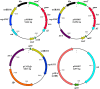
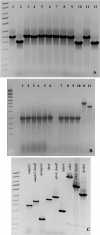
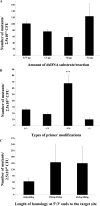
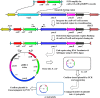
The genetic modification of primary bacterial disease isolates is challenging due to the lack of highly efficient genetic tools. Herein we describe the development of a modified PCR-based, λ Red-mediated recombineering system for efficient deletion of genes in Gram-negative bacteria. A series of conjugally transferrable plasmids were constructed by cloning an oriT sequence and different antibiotic resistance genes into recombinogenic plasmid pKD46. Using this system we deleted ten different genes from the genomes of Edwardsiella ictaluri and Aeromonas hydrophila. A temperature sensitive and conjugally transferable flp recombinase plasmid was developed to generate markerless gene deletion mutants. We also developed an efficient cloning system to capture larger bacterial genetic elements and clone them into a conjugally transferrable plasmid for facile transferring to Gram-negative bacteria. This system should be applicable in diverse Gram-negative bacteria to modify and complement genomic elements in bacteria that cannot be manipulated using available genetic tools.
Keywords: Bacterial pathogens; Genetic modification; Recombineering.
Figures
References
-
- Ando T., Xu Q., Torres M., Kusugami K., Israel D.A., Blaser M.J. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 2000;37:1052–1065. - PubMed
-
- Arber W., Dussoix D. Host specificity of DNA produced by E. coli I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 1962;5:18–36. - PubMed
-
- Arber W., Linn S. DNA modification and restriction. Annu. Rev. Biochem. 1969;38:467–500. - PubMed
-
- Aubert D.F., Hamad M.A., Valvano M.A. A markerless deletion method for genetic manipulation of Burkholderia cenocepacia and other multidrug-resistant gram-negative bacteria. Methods Mol. Biol. 2014;1197:311–327. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
