Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore
- PMID: 28353339
- PMCID: PMC5444049
- DOI: 10.1021/acsnano.6b07760
Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore
Abstract
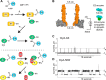
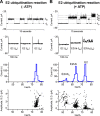
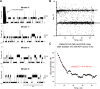
The covalent addition of ubiquitin to target proteins is a key post-translational modification that is linked to a myriad of biological processes. Here, we report a fast, single-molecule, and label-free method to probe the ubiquitination of proteins employing an engineered Cytolysin A (ClyA) nanopore. We show that ionic currents can be used to recognize mono- and polyubiquitinated forms of native proteins under physiological conditions. Using defined conjugates, we also show that isomeric monoubiquitinated proteins can be discriminated. The nanopore approach allows following the ubiquitination reaction in real time, which will accelerate the understanding of fundamental mechanisms linked to protein ubiquitination.
Keywords: nanopore; nanotechnology; protein modifications; single-molecule kinetics; ubiquitin.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Hershko A.; Heller H.; Elias S.; Ciechanover A. Components of Ubiquitin-Protein Ligase System. Resolution, Affinity Purification, and Role in Protein Breakdown. J. Biol. Chem. 1983, 258, 8206–8214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

