Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis
- PMID: 28355185
- PMCID: PMC5383507
- DOI: 10.1038/nature21693
Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis
Abstract
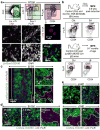
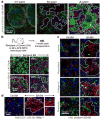
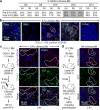
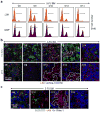
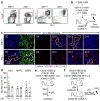
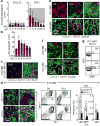
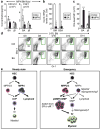
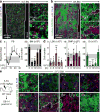
Although many aspects of blood production are well understood, the spatial organization of myeloid differentiation in the bone marrow remains unknown. Here we use imaging to track granulocyte/macrophage progenitor (GMP) behaviour in mice during emergency and leukaemic myelopoiesis. In the steady state, we find individual GMPs scattered throughout the bone marrow. During regeneration, we observe expanding GMP patches forming defined GMP clusters, which, in turn, locally differentiate into granulocytes. The timed release of important bone marrow niche signals (SCF, IL-1β, G-CSF, TGFβ and CXCL4) and activation of an inducible Irf8 and β-catenin progenitor self-renewal network control the transient formation of regenerating GMP clusters. In leukaemia, we show that GMP clusters are constantly produced owing to persistent activation of the self-renewal network and a lack of termination cytokines that normally restore haematopoietic stem-cell quiescence. Our results uncover a previously unrecognized dynamic behaviour of GMPs in situ, which tunes emergency myelopoiesis and is hijacked in leukaemia.
Conflict of interest statement
Figures



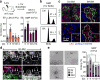

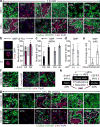
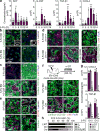

Comment in
-
GMP-ing to Spatial Conclusions about Emergency and Leukemic Myelopoiesis.Cell Stem Cell. 2017 May 4;20(5):579-581. doi: 10.1016/j.stem.2017.04.005. Cell Stem Cell. 2017. PMID: 28475879 Free PMC article.
References
-
- Busch K, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. - PubMed
-
- Cabezas-Wallscheid N, et al. Identification of regulatory networks in HSC and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–522. - PubMed
-
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

