Precise Temporal Profiling of Signaling Complexes in Primary Cells Using SWATH Mass Spectrometry
- PMID: 28355572
- PMCID: PMC5382234
- DOI: 10.1016/j.celrep.2017.03.019
Precise Temporal Profiling of Signaling Complexes in Primary Cells Using SWATH Mass Spectrometry
Abstract
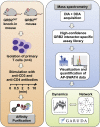
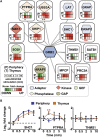
Spatiotemporal organization of protein interactions in cell signaling is a fundamental process that drives cellular functions. Given differential protein expression across tissues and developmental stages, the architecture and dynamics of signaling interaction proteomes is, likely, highly context dependent. However, current interaction information has been almost exclusively obtained from transformed cells. In this study, we applied an advanced and robust workflow combining mouse genetics and affinity purification (AP)-SWATH mass spectrometry to profile the dynamics of 53 high-confidence protein interactions in primary T cells, using the scaffold protein GRB2 as a model. The workflow also provided a sufficient level of robustness to pinpoint differential interaction dynamics between two similar, but functionally distinct, primary T cell populations. Altogether, we demonstrated that precise and reproducible quantitative measurements of protein interaction dynamics can be achieved in primary cells isolated from mammalian tissues, allowing resolution of the tissue-specific context of cell-signaling events.
Keywords: DIA; GRB2; SWATH; interactome; primary T cells; targeted mass spectrometry.
Crown Copyright © 2017. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. - PubMed
-
- Astoul E., Edmunds C., Cantrell D.A., Ward S.G. PI 3-K and T-cell activation: limitations of T-leukemic cell lines as signaling models. Trends Immunol. 2001;22:490–496. - PubMed
-
- Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B., Vitek O. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. - PubMed
-
- Collins B.C., Gillet L.C., Rosenberger G., Röst H.L., Vichalkovski A., Gstaiger M., Aebersold R. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods. 2013;10:1246–1253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

