Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles
- PMID: 28355876
- PMCID: PMC5898444
- DOI: 10.1021/jacs.7b01791
Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles
Abstract
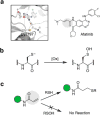
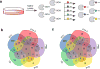
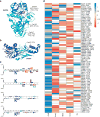
Targeted covalent inhibitors have emerged as a powerful approach in the drug discovery pipeline. Key to this process is the identification of signaling pathways (or receptors) specific to (or overexpressed in) disease cells. In this context, fragment-based ligand discovery (FBLD) has significantly expanded our view of the ligandable proteome and affords tool compounds for biological inquiry. To date, such covalent ligand discovery has almost exclusively employed cysteine-reactive small-molecule fragments. However, functional cysteine residues in proteins are often redox-sensitive and can undergo oxidation in cells. Such reactions are particularly relevant in diseases, like cancer, which are linked to excessive production of reactive oxygen species. Once oxidized, the sulfur atom of cysteine is much less reactive toward electrophilic groups used in the traditional FBLD paradigm. To address this limitation, we recently developed a novel library of diverse carbon-based nucleophile fragments that react selectively with cysteine sulfenic acid formed in proteins via oxidation or hydrolysis reactions. Here, we report analysis of sulfenic acid-reactive C-nucleophile fragments screened against a colon cancer cell proteome. Covalent ligands were identified for >1280 S-sulfenylated cysteines present in "druggable" proteins and orphan targets, revealing disparate reactivity profiles and target preferences. Among the unique ligand-protein interactions identified was that of a pyrrolidinedione nucleophile that reacted preferentially with protein tyrosine phosphatases. Fragment-based covalent ligand discovery with C-nucleophiles affords an expansive snapshot of the ligandable "redoxome" with significant implications for covalent inhibitor pharmacology and also affords new chemical tools to investigate redox-regulation of protein function.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
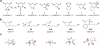

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

