Core-genome scaffold comparison reveals the prevalence that inversion events are associated with pairs of inverted repeats
- PMID: 28356070
- PMCID: PMC5372343
- DOI: 10.1186/s12864-017-3655-0
Core-genome scaffold comparison reveals the prevalence that inversion events are associated with pairs of inverted repeats
Abstract
Background: Genome rearrangement describes gross changes of chromosomal regions, plays an important role in evolutionary biology and has profound impacts on phenotype in organisms ranging from microbes to humans. With more and more complete genomes accomplished, lots of genomic comparisons have been conducted in order to find genome rearrangements and the mechanisms which underlie the rearrangement events. In our opinion, genomic comparison of different individuals/strains within the same species (pan-genome) is more helpful to reveal the mechanisms for genome rearrangements since genomes of the same species are much closer to each other.
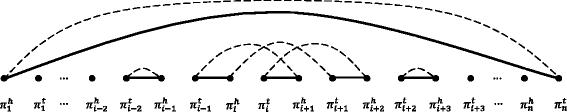
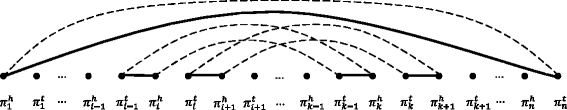
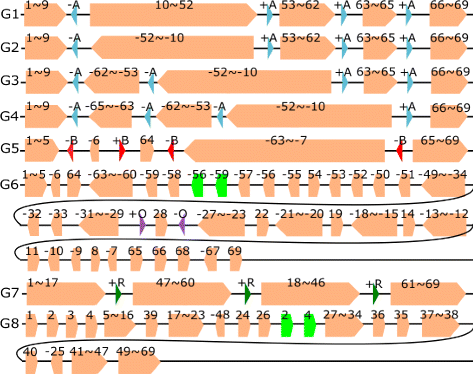
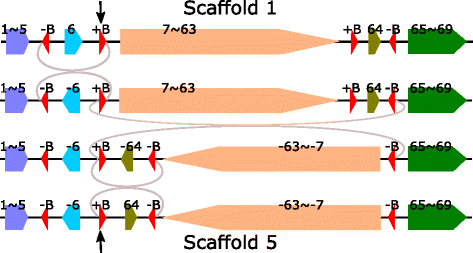
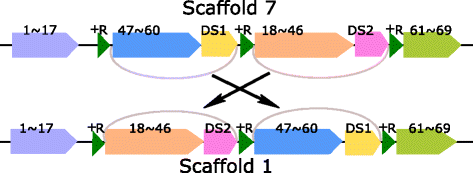
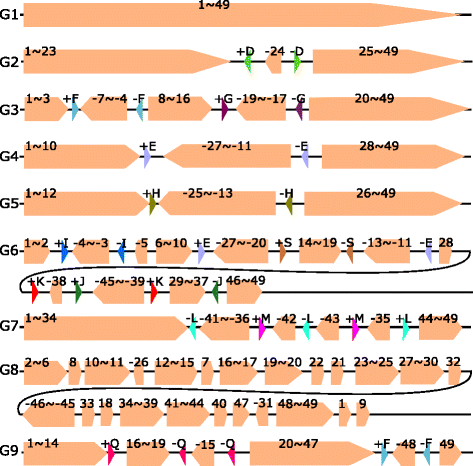
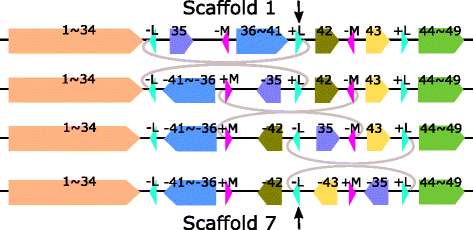
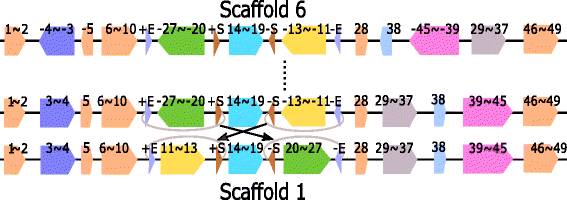
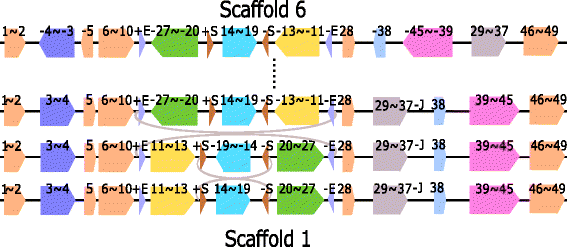
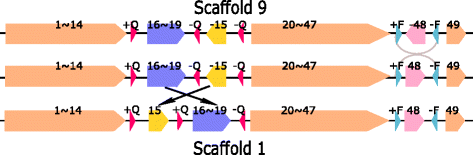
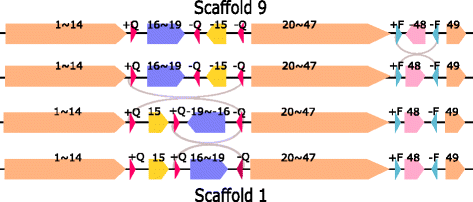
Results: We study the mechanism for inversion events via core-genome scaffold comparison of different strains within the same species. We focus on two kinds of bacteria, Pseudomonas aeruginosa and Escherichia coli, and investigate the inversion events among different strains of the same species. We find an interesting phenomenon that long (larger than 10,000 bp) inversion regions are flanked by a pair of Inverted Repeats (IRs). This mechanism can also explain why the breakpoint reuses for inversion events happen. We study the prevalence of the phenomenon and find that it is a major mechanism for inversions. The other observation is that for different rearrangement events such as transposition and inverted block interchange, the two ends of the swapped regions are also associated with repeats so that after the rearrangement operations the two ends of the swapped regions remain unchanged. To our knowledge, this is the first time such a phenomenon is reported for transposition event.
Conclusions: In both Pseudomonas aeruginosa and Escherichia coli strains, IRs were found at the two ends of long sequence inversions. The two ends of the inversion remained unchanged before and after the inversion event. The existence of IRs can explain the breakpoint reuse phenomenon. We also observed that other rearrangement operations such as transposition, inverted transposition, and inverted block interchange, had repeats (not necessarily inverted) at the ends of each segment, where the ends remained unchanged before and after the rearrangement operations. This suggests that the conservation of ends could possibly be a popular phenomenon in many types of chromosome rearrangement events.
Keywords: Comparative genomics; Genome rearrangment; Inversion; Inverted block interchange; Transposition.
Figures
Similar articles
-
GRSR: a tool for deriving genome rearrangement scenarios from multiple unichromosomal genome sequences.BMC Bioinformatics. 2018 Aug 13;19(Suppl 9):291. doi: 10.1186/s12859-018-2268-1. BMC Bioinformatics. 2018. PMID: 30367596 Free PMC article.
-
DNA rearrangement mediated by inverted repeats.Proc Natl Acad Sci U S A. 1996 Jan 23;93(2):819-23. doi: 10.1073/pnas.93.2.819. Proc Natl Acad Sci U S A. 1996. PMID: 8570641 Free PMC article.
-
DNA inversions between short inverted repeats in Escherichia coli.Genetics. 1992 Oct;132(2):295-302. doi: 10.1093/genetics/132.2.295. Genetics. 1992. PMID: 1427029 Free PMC article.
-
Sorting by weighted reversals, transpositions, and inverted transpositions.J Comput Biol. 2007 Jun;14(5):615-36. doi: 10.1089/cmb.2007.R006. J Comput Biol. 2007. PMID: 17683264 Review.
-
Repeated sequences in bacterial chromosomes and plasmids: a glimpse from sequenced genomes.Res Microbiol. 1999 Nov-Dec;150(9-10):735-43. doi: 10.1016/s0923-2508(99)00119-9. Res Microbiol. 1999. PMID: 10673011 Review.
Cited by
-
Micro-evolution of three Streptococcus species: selection, antigenic variation, and horizontal gene inflow.BMC Evol Biol. 2019 Mar 27;19(1):83. doi: 10.1186/s12862-019-1403-6. BMC Evol Biol. 2019. PMID: 30917781 Free PMC article.
-
Genetic Markers of Genome Rearrangements in Helicobacter pylori.Microorganisms. 2021 Mar 17;9(3):621. doi: 10.3390/microorganisms9030621. Microorganisms. 2021. PMID: 33802974 Free PMC article.
-
Positive Selection during Niche Adaptation Results in Large-Scale and Irreversible Rearrangement of Chromosomal Gene Order in Bacteria.Mol Biol Evol. 2022 Apr 10;39(4):msac069. doi: 10.1093/molbev/msac069. Mol Biol Evol. 2022. PMID: 35348727 Free PMC article.
-
MetaPGN: a pipeline for construction and graphical visualization of annotated pangenome networks.Gigascience. 2018 Nov 1;7(11):giy121. doi: 10.1093/gigascience/giy121. Gigascience. 2018. PMID: 30277499 Free PMC article.
-
Chronological set of E. coli O157:H7 bovine strains establishes a role for repeat sequences and mobile genetic elements in genome diversification.BMC Genomics. 2020 Aug 17;21(1):562. doi: 10.1186/s12864-020-06943-x. BMC Genomics. 2020. PMID: 32807088 Free PMC article.
References
-
- Li Z, Wang L, Zhang K. Algorithmic approaches for genome rearrangement: a review. IEEE Trans Syst Man Cybern Part C Appl Rev. 2006;36(5):636–48. doi: 10.1109/TSMCC.2005.855522. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

