CNV discovery for milk composition traits in dairy cattle using whole genome resequencing
- PMID: 28356085
- PMCID: PMC5371188
- DOI: 10.1186/s12864-017-3636-3
CNV discovery for milk composition traits in dairy cattle using whole genome resequencing
Abstract
Background: Copy number variations (CNVs) are important and widely distributed in the genome. CNV detection opens a new avenue for exploring genes associated with complex traits in humans, animals and plants. Herein, we present a genome-wide assessment of CNVs that are potentially associated with milk composition traits in dairy cattle.
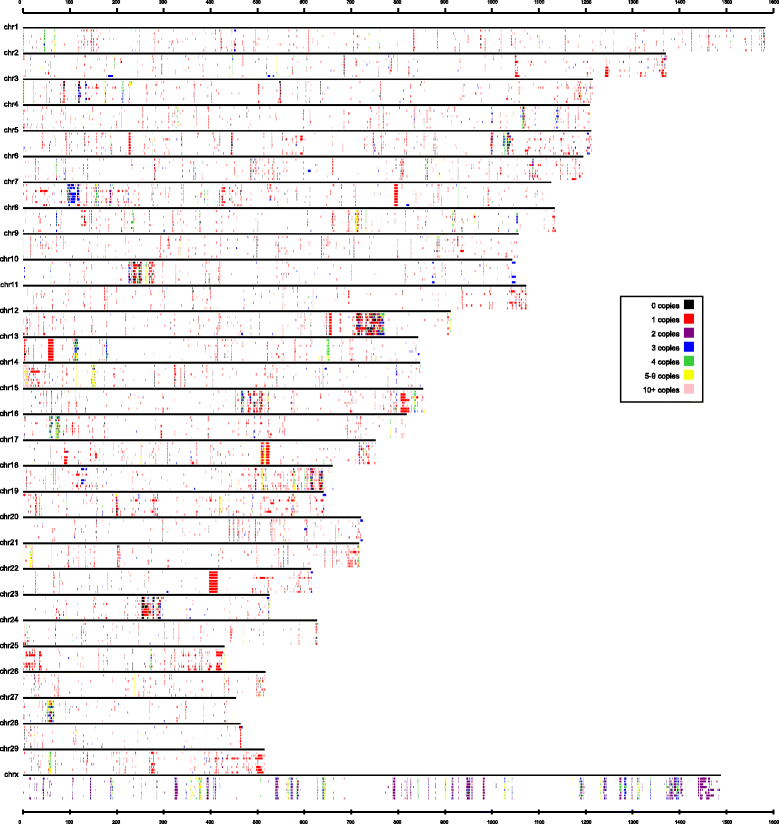
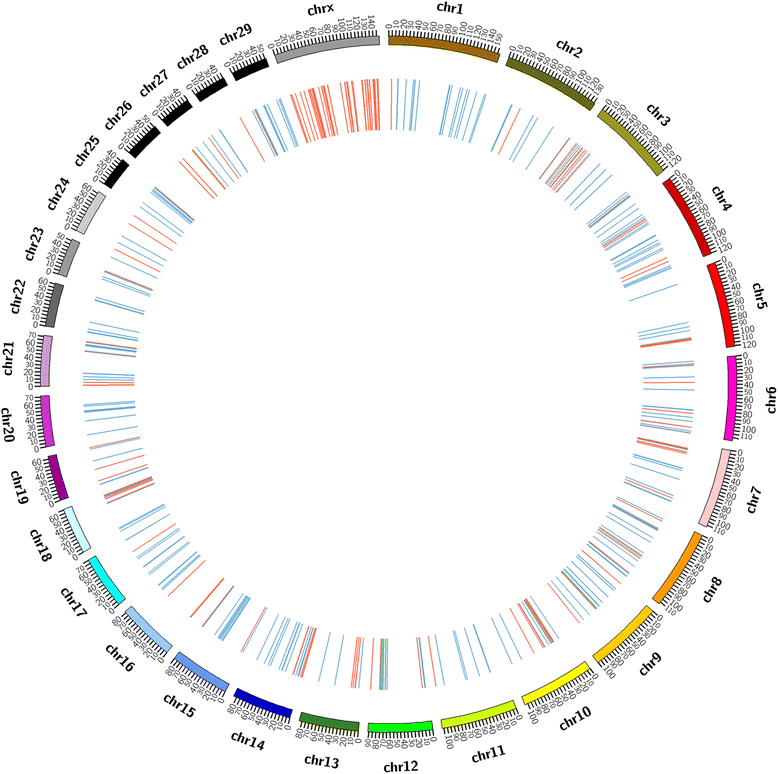
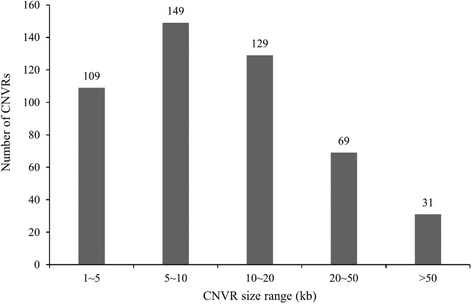
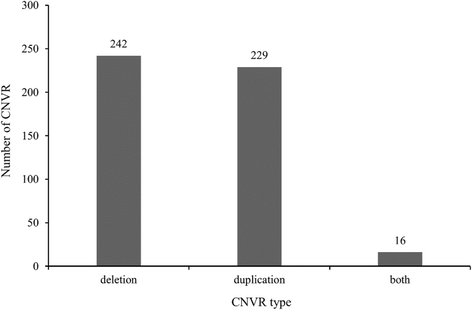
Results: In this study, CNVs were detected based on whole genome re-sequencing data of eight Holstein bulls from four half- and/or full-sib families, with extremely high and low estimated breeding values (EBVs) of milk protein percentage and fat percentage. The range of coverage depth per individual was 8.2-11.9×. Using CNVnator, we identified a total of 14,821 CNVs, including 5025 duplications and 9796 deletions. Among them, 487 differential CNV regions (CNVRs) comprising ~8.23 Mb of the cattle genome were observed between the high and low groups. Annotation of these differential CNVRs were performed based on the cattle genome reference assembly (UMD3.1) and totally 235 functional genes were found within the CNVRs. By Gene Ontology and KEGG pathway analyses, we found that genes were significantly enriched for specific biological functions related to protein and lipid metabolism, insulin/IGF pathway-protein kinase B signaling cascade, prolactin signaling pathway and AMPK signaling pathways. These genes included INS, IGF2, FOXO3, TH, SCD5, GALNT18, GALNT16, ART3, SNCA and WNT7A, implying their potential association with milk protein and fat traits. In addition, 95 CNVRs were overlapped with 75 known QTLs that are associated with milk protein and fat traits of dairy cattle (Cattle QTLdb).
Conclusions: In conclusion, based on NGS of 8 Holstein bulls with extremely high and low EBVs for milk PP and FP, we identified a total of 14,821 CNVs, 487 differential CNVRs between groups, and 10 genes, which were suggested as promising candidate genes for milk protein and fat traits.
Keywords: Chinese Holstein; Copy number variation; Whole genome re-sequencing.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

