Integrative clustering reveals a novel split in the luminal A subtype of breast cancer with impact on outcome
- PMID: 28356166
- PMCID: PMC5372339
- DOI: 10.1186/s13058-017-0812-y
Integrative clustering reveals a novel split in the luminal A subtype of breast cancer with impact on outcome
Abstract
Background: Breast cancer is a heterogeneous disease at the clinical and molecular level. In this study we integrate classifications extracted from five different molecular levels in order to identify integrated subtypes.
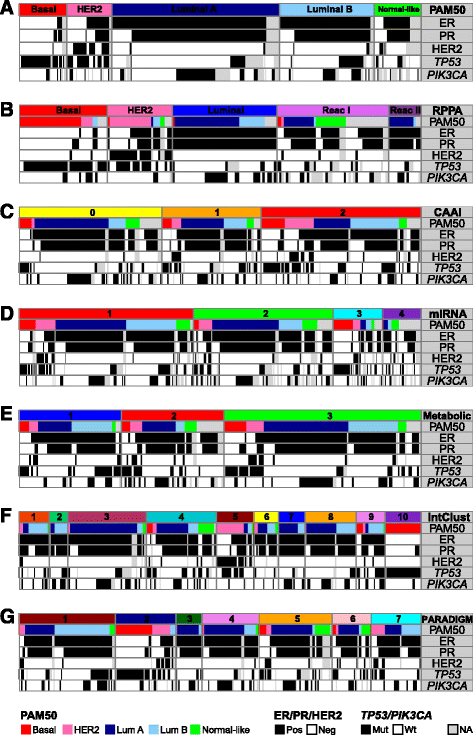
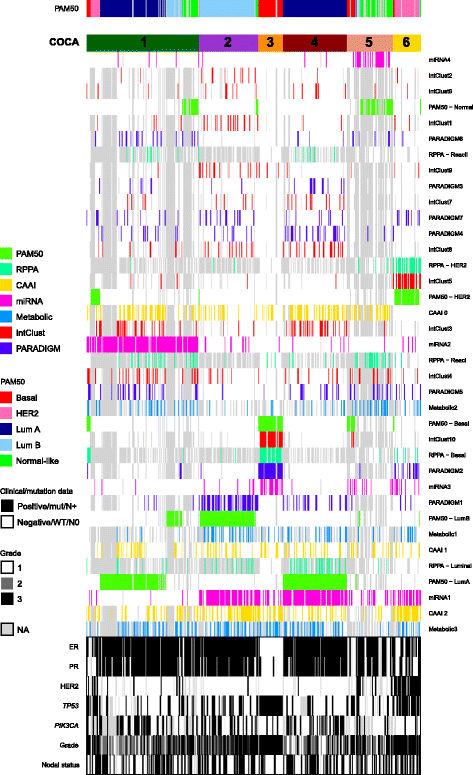
Methods: Tumor tissue from 425 patients with primary breast cancer from the Oslo2 study was cut and blended, and divided into fractions for DNA, RNA and protein isolation and metabolomics, allowing the acquisition of representative and comparable molecular data. Patients were stratified into groups based on their tumor characteristics from five different molecular levels, using various clustering methods. Finally, all previously identified and newly determined subgroups were combined in a multilevel classification using a "cluster-of-clusters" approach with consensus clustering.
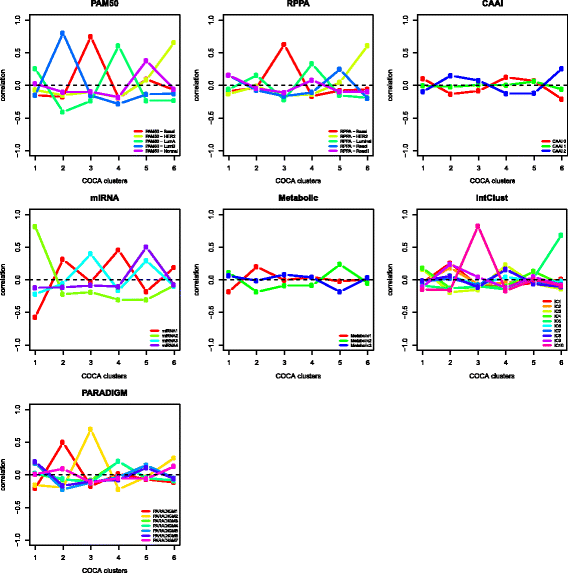
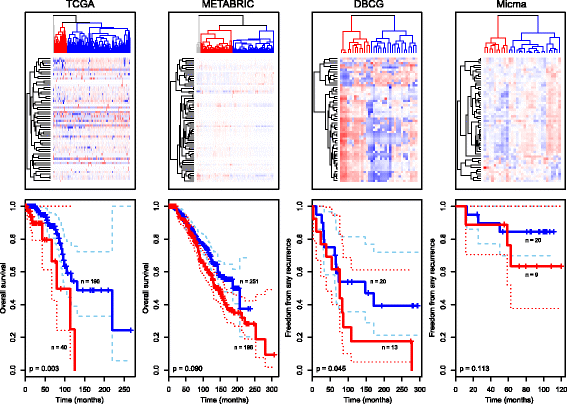
Results: Based on DNA copy number data, tumors were categorized into three groups according to the complex arm aberration index. mRNA expression profiles divided tumors into five molecular subgroups according to PAM50 subtyping, and clustering based on microRNA expression revealed four subgroups. Reverse-phase protein array data divided tumors into five subgroups. Hierarchical clustering of tumor metabolic profiles revealed three clusters. Combining DNA copy number and mRNA expression classified tumors into seven clusters based on pathway activity levels, and tumors were classified into ten subtypes using integrative clustering. The final consensus clustering that incorporated all aforementioned subtypes revealed six major groups. Five corresponded well with the mRNA subtypes, while a sixth group resulted from a split of the luminal A subtype; these tumors belonged to distinct microRNA clusters. Gain-of-function studies using MCF-7 cells showed that microRNAs differentially expressed between the luminal A clusters were important for cancer cell survival. These microRNAs were used to validate the split in luminal A tumors in four independent breast cancer cohorts. In two cohorts the microRNAs divided tumors into subgroups with significantly different outcomes, and in another a trend was observed.
Conclusions: The six integrated subtypes identified confirm the heterogeneity of breast cancer and show that finer subdivisions of subtypes are evident. Increasing knowledge of the heterogeneity of the luminal A subtype may add pivotal information to guide therapeutic choices, evidently bringing us closer to improved treatment for this largest subgroup of breast cancer.
Keywords: Breast cancer; Consensus clustering; Integration; Luminal A; MicroRNA.
Figures
References
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
-
- Kristensen VN, Vaske CJ, Ursini-Siegel J, Van Loo P, Nordgard SH, Sachidanandam R, Sørlie T, Wärnberg F, Haakensen VD, Helland Å, et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci. 2012;109(8):2802–7. doi: 10.1073/pnas.1108781108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

