The crossover conformational shift of the GTPase atlastin provides the energy driving ER fusion
- PMID: 28356327
- PMCID: PMC5412568
- DOI: 10.1083/jcb.201609071
The crossover conformational shift of the GTPase atlastin provides the energy driving ER fusion
Abstract
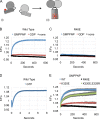
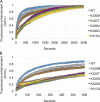
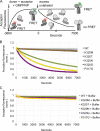
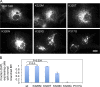
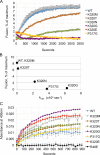
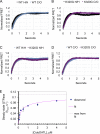
The homotypic fusion of endoplasmic reticulum membranes is catalyzed by the atlastin GTPase. The mechanism involves trans-dimerization between GTPase heads and a favorable crossover conformational shift, catalyzed by GTP hydrolysis, that converts the dimer from a "prefusion" to "postfusion" state. However, whether crossover formation actually energizes fusion remains unclear, as do the sequence of events surrounding it. Here, we made mutations in atlastin to selectively destabilize the crossover conformation and used fluorescence-based kinetic assays to analyze the variants. All variants underwent dimerization and crossover concurrently, and at wild-type rates. However, certain variants were unstable once in the crossover dimer conformation, and crossover dimer stability closely paralleled lipid-mixing activity. Tethering, however, appeared to be unimpaired in all mutant variants. The results suggest that tethering and lipid mixing are catalyzed concurrently by GTP hydrolysis but that the energy requirement for lipid mixing exceeds that for tethering, and the full energy released through crossover formation is necessary for fusion.
© 2017 Winsor et al.
Figures




References
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

