Rearrangement of a polar core provides a conserved mechanism for constitutive activation of class B G protein-coupled receptors
- PMID: 28356352
- PMCID: PMC5473240
- DOI: 10.1074/jbc.M117.782987
Rearrangement of a polar core provides a conserved mechanism for constitutive activation of class B G protein-coupled receptors
Abstract
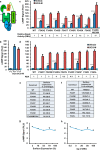
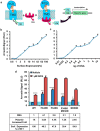
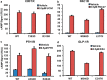
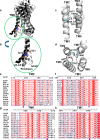
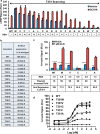
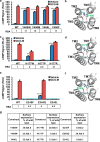
The glucagon receptor (GCGR) belongs to the secretin-like (class B) family of G protein-coupled receptors (GPCRs) and is activated by the peptide hormone glucagon. The structures of an activated class B GPCR have remained unsolved, preventing a mechanistic understanding of how these receptors are activated. Using a combination of structural modeling and mutagenesis studies, we present here two modes of ligand-independent activation of GCGR. First, we identified a GCGR-specific hydrophobic lock comprising Met-338 and Phe-345 within the IC3 loop and transmembrane helix 6 (TM6) and found that this lock stabilizes the TM6 helix in the inactive conformation. Disruption of this hydrophobic lock led to constitutive G protein and arrestin signaling. Second, we discovered a polar core comprising conserved residues in TM2, TM3, TM6, and TM7, and mutations that disrupt this polar core led to constitutive GCGR activity. On the basis of these results, we propose a mechanistic model of GCGR activation in which TM6 is held in an inactive conformation by the conserved polar core and the hydrophobic lock. Mutations that disrupt these inhibitory elements allow TM6 to swing outward to adopt an active TM6 conformation similar to that of the canonical β2-adrenergic receptor complexed with G protein and to that of rhodopsin complexed with arrestin. Importantly, mutations in the corresponding polar core of several other members of class B GPCRs, including PTH1R, PAC1R, VIP1R, and CRFR1, also induce constitutive G protein signaling, suggesting that the rearrangement of the polar core is a conserved mechanism for class B GPCR activation.
Keywords: 7-helix receptor; G protein-coupled receptor (GPCR); arrestin; glucagon; parathyroid hormone.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
-
- Lagerström M. C., and Schiöth H. B. (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 - PubMed
-
- Hoare S. R. (2005) Mechanisms of peptide and nonpeptide ligand binding to Class B G-protein-coupled receptors. Drug Discov. Today 10, 417–427 - PubMed
-
- Parthier C., Reedtz-Runge S., Rudolph R., and Stubbs M. T. (2009) Passing the baton in class B GPCRs: peptide hormone activation via helix induction? Trends Biochem. Sci. 34, 303–310 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

