The druggable genome and support for target identification and validation in drug development
- PMID: 28356508
- PMCID: PMC6321762
- DOI: 10.1126/scitranslmed.aag1166
The druggable genome and support for target identification and validation in drug development
Abstract
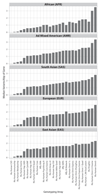
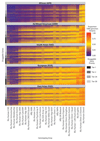
Target identification (determining the correct drug targets for a disease) and target validation (demonstrating an effect of target perturbation on disease biomarkers and disease end points) are important steps in drug development. Clinically relevant associations of variants in genes encoding drug targets model the effect of modifying the same targets pharmacologically. To delineate drug development (including repurposing) opportunities arising from this paradigm, we connected complex disease- and biomarker-associated loci from genome-wide association studies to an updated set of genes encoding druggable human proteins, to agents with bioactivity against these targets, and, where there were licensed drugs, to clinical indications. We used this set of genes to inform the design of a new genotyping array, which will enable association studies of druggable genes for drug target selection and validation in human disease.
Copyright © 2017, American Association for the Advancement of Science.
Conflict of interest statement
Figures


References
-
- Munos B. Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov. 2009;8:959–968. - PubMed
-
- Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203–214. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

