Minute Virus of Canines NP1 Protein Governs the Expression of a Subset of Essential Nonstructural Proteins via Its Role in RNA Processing
- PMID: 28356522
- PMCID: PMC5446662
- DOI: 10.1128/JVI.00260-17
Minute Virus of Canines NP1 Protein Governs the Expression of a Subset of Essential Nonstructural Proteins via Its Role in RNA Processing
Abstract
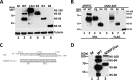
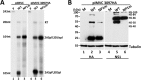
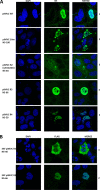
Parvoviruses use a variety of means to control the expression of their compact genomes. The bocaparvovirus minute virus of canines (MVC) encodes a small, genus-specific protein, NP1, which governs access to the viral capsid gene via its role in alternative polyadenylation and alternative splicing of the single MVC pre-mRNA. In addition to NP1, MVC encodes five additional nonstructural proteins (NS) that share an initiation codon at the left end of the genome and which are individually encoded by alternative multiply spliced mRNAs. We found that three of these proteins were encoded by mRNAs that excise the NP1-regulated MVC intron immediately upstream of the internal polyadenylation site, (pA)p, and that generation of these proteins was thus regulated by NP1. Splicing of their progenitor mRNAs joined the amino termini of these proteins to the NP1 open reading frame, and splice site mutations that prevented their expression inhibited virus replication in a host cell-dependent manner. Thus, in addition to controlling capsid gene access, NP1 also controls the expression of three of the five identified NS proteins via its role in governing MVC pre-mRNA splicing.IMPORTANCE The Parvovirinae are small nonenveloped icosahedral viruses that are important pathogens in many animal species, including humans. Minute virus of canine (MVC) is an autonomous parvovirus in the genus Bocaparvovirus It has a single promoter that generates a single pre-mRNA. NP1, a small genus-specific MVC protein, participates in the processing of this pre-mRNA and so controls capsid gene access via its role in alternative internal polyadenylation and splicing. We show that NP1 also controls the expression of three of the five identified NS proteins via its role in governing MVC pre-mRNA splicing. These NS proteins together are required for virus replication in a host cell-dependent manner.
Keywords: gene expression; parvovirus.
Copyright © 2017 American Society for Microbiology.
Figures


Similar articles
-
Parvovirus RNA Processing: Compact Genomic Organization and Unique Alternative mRNA Processing Mechanisms.Viruses. 2025 Jul 15;17(7):984. doi: 10.3390/v17070984. Viruses. 2025. PMID: 40733601 Free PMC article. Review.
-
NP1 Protein of the Bocaparvovirus Minute Virus of Canines Controls Access to the Viral Capsid Genes via Its Role in RNA Processing.J Virol. 2015 Dec 4;90(4):1718-28. doi: 10.1128/JVI.02618-15. Print 2016 Feb 15. J Virol. 2015. PMID: 26637456 Free PMC article.
-
The Human Bocavirus 1 NP1 Protein Is a Multifunctional Regulator of Viral RNA Processing.J Virol. 2018 Oct 29;92(22):e01187-18. doi: 10.1128/JVI.01187-18. Print 2018 Nov 15. J Virol. 2018. PMID: 30135129 Free PMC article.
-
Minute Virus of Canines NP1 Protein Interacts with the Cellular Factor CPSF6 To Regulate Viral Alternative RNA Processing.J Virol. 2019 Jan 4;93(2):e01530-18. doi: 10.1128/JVI.01530-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30355695 Free PMC article.
-
Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
Cited by
-
Recent Advances in Molecular Biology of Human Bocavirus 1 and Its Applications.Front Microbiol. 2021 Jun 16;12:696604. doi: 10.3389/fmicb.2021.696604. eCollection 2021. Front Microbiol. 2021. PMID: 34220786 Free PMC article. Review.
-
The Minute Virus of Canines (MVC) Activates the RhoA/ROCK1/MLC2 Signal Transduction Pathway Resulting in the Dissociation of Tight Junctions and Facilitating Occludin-Mediated Viral Infection.Microorganisms. 2025 Mar 20;13(3):695. doi: 10.3390/microorganisms13030695. Microorganisms. 2025. PMID: 40142587 Free PMC article.
-
Parvovirus RNA Processing: Compact Genomic Organization and Unique Alternative mRNA Processing Mechanisms.Viruses. 2025 Jul 15;17(7):984. doi: 10.3390/v17070984. Viruses. 2025. PMID: 40733601 Free PMC article. Review.
-
Cellular Cleavage and Polyadenylation Specificity Factor 6 (CPSF6) Mediates Nuclear Import of Human Bocavirus 1 NP1 Protein and Modulates Viral Capsid Protein Expression.J Virol. 2020 Jan 6;94(2):e01444-19. doi: 10.1128/JVI.01444-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666379 Free PMC article.
-
N4-acetylcytidine coordinates with NP1 and CPSF5 to facilitate alternative RNA processing during the replication of minute virus of canines.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf229. doi: 10.1093/nar/gkaf229. Nucleic Acids Res. 2025. PMID: 40167508 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

