A single mutation in the 15S rRNA gene confers non sense suppressor activity and interacts with mRF1 the release factor in yeast mitochondria
- PMID: 28357310
- PMCID: PMC5354577
- DOI: 10.15698/mic2015.09.223
A single mutation in the 15S rRNA gene confers non sense suppressor activity and interacts with mRF1 the release factor in yeast mitochondria
Abstract
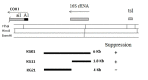
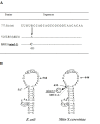
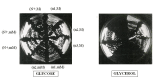
We have determined the nucleotide sequence of the mim3-1 mitochondrial ribosomal suppressor, acting on ochre mitochondrial mutations and one frameshift mutation in Saccharomyces cerevisiae. The 15s rRNA suppressor gene contains a G633 to C transversion. Yeast mitochondrial G633 corresponds to G517 of the E.coli 15S rRNA, which is occupied by an invariant G in all known small rRNA sequences. Interestingly, this mutation has occurred at the same position as the known MSU1 mitochondrial suppressor which changes G633 to A. The suppressor mutation lies in a highly conserved region of the rRNA, known in E.coli as the 530-loop, interacting with the S4, S5 and S12 ribosomal proteins. We also show an interesting interaction between the mitochondrial mim3-1 and the nuclear nam3-1 suppressors, both of which have the same action spectrum on mitochondrial mutations: nam3-1 abolishes the suppressor effect when present with mim3-1 in the same haploid cell. We discuss these results in the light of the nature of Nam3, identified by 1 as the yeast mitochondrial translation release factor. A hypothetical mechanism of suppression by "ribosome shifting" is also discussed in view of the nature of mutations suppressed and not suppressed.
Keywords: 15S rRNA; frame shift; informational suppressors; nonsense; yeast.
Conflict of interest statement
Conflict of interest: All the authors declare no conflict of interest.
Figures

Similar articles
-
Substitution of an invariant nucleotide at the base of the highly conserved '530-loop' of 15S rRNA causes suppression of yeast mitochondrial ochre mutations.Nucleic Acids Res. 1989 Jun 26;17(12):4535-9. doi: 10.1093/nar/17.12.4535. Nucleic Acids Res. 1989. PMID: 2473436 Free PMC article.
-
mim3 and nam3 omnipotent suppressor genes similarly affect the polypeptide composition of yeast mitoribosomes.Biochimie. 1987 May;69(5):531-7. doi: 10.1016/0300-9084(87)90090-3. Biochimie. 1987. PMID: 3118968
-
Suppressor of yeast mitochondrial ochre mutations that maps in or near the 15S ribosomal RNA gene of mtDNA.Proc Natl Acad Sci U S A. 1982 Mar;79(5):1583-7. doi: 10.1073/pnas.79.5.1583. Proc Natl Acad Sci U S A. 1982. PMID: 6280192 Free PMC article.
-
Polypeptide chain termination in Saccharomyces cerevisiae.Curr Genet. 1994 May;25(5):385-95. doi: 10.1007/BF00351776. Curr Genet. 1994. PMID: 8082183 Review.
-
Modulation of efficiency of translation termination in Saccharomyces cerevisiae.Prion. 2014;8(3):247-60. doi: 10.4161/pri.29851. Epub 2014 Nov 1. Prion. 2014. PMID: 25486049 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
