Functions and regulation of the MRX complex at DNA double-strand breaks
- PMID: 28357369
- PMCID: PMC5349012
- DOI: 10.15698/mic2016.08.517
Functions and regulation of the MRX complex at DNA double-strand breaks
Abstract
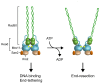
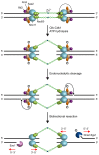
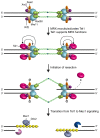
DNA double-strand breaks (DSBs) pose a serious threat to genome stability and cell survival. Cells possess mechanisms that recognize DSBs and promote their repair through either homologous recombination (HR) or non-homologous end joining (NHEJ). The evolutionarily conserved Mre11-Rad50-Xrs2 (MRX) complex plays a central role in the cellular response to DSBs, as it is implicated in controlling end resection and in maintaining the DSB ends tethered to each other. Furthermore, it is responsible for DSB signaling by activating the checkpoint kinase Tel1 that, in turn, supports MRX function in a positive feedback loop. The present review focuses mainly on recent works in the budding yeast Saccharomyces cerevisiae to highlight structure and regulation of MRX as well as its interplays with Tel1.
Keywords: MRX; Rif2; Sae2; Tel1; double-strand break; nucleases; resection.
Conflict of interest statement
Conflict of interest: The authors declare no conflict of interest.
Figures
Similar articles
-
Tel1 and Rif2 Regulate MRX Functions in End-Tethering and Repair of DNA Double-Strand Breaks.PLoS Biol. 2016 Feb 22;14(2):e1002387. doi: 10.1371/journal.pbio.1002387. eCollection 2016 Feb. PLoS Biol. 2016. PMID: 26901759 Free PMC article.
-
Interplay between Sae2 and Rif2 in the regulation of Mre11-Rad50 activities at DNA ends.Curr Opin Genet Dev. 2021 Dec;71:72-77. doi: 10.1016/j.gde.2021.07.001. Epub 2021 Jul 24. Curr Opin Genet Dev. 2021. PMID: 34311383 Review.
-
Functional and structural insights into the MRX/MRN complex, a key player in recognition and repair of DNA double-strand breaks.Comput Struct Biotechnol J. 2020 May 16;18:1137-1152. doi: 10.1016/j.csbj.2020.05.013. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32489527 Free PMC article. Review.
-
Processing of DNA Double-Strand Breaks by the MRX Complex in a Chromatin Context.Front Mol Biosci. 2019 Jun 7;6:43. doi: 10.3389/fmolb.2019.00043. eCollection 2019. Front Mol Biosci. 2019. PMID: 31231660 Free PMC article. Review.
-
The MRX Complex Ensures NHEJ Fidelity through Multiple Pathways Including Xrs2-FHA-Dependent Tel1 Activation.PLoS Genet. 2016 Mar 18;12(3):e1005942. doi: 10.1371/journal.pgen.1005942. eCollection 2016 Mar. PLoS Genet. 2016. PMID: 26990569 Free PMC article.
Cited by
-
DNA binding modes influence Rap1 activity in the regulation of telomere length and MRX functions at DNA ends.Nucleic Acids Res. 2020 Mar 18;48(5):2424-2441. doi: 10.1093/nar/gkz1203. Nucleic Acids Res. 2020. PMID: 31879780 Free PMC article.
-
Recruitment kinetics of the homologous recombination pathway in procyclic forms of Trypanosoma brucei after ionizing radiation treatment.Sci Rep. 2018 Mar 29;8(1):5405. doi: 10.1038/s41598-018-23731-6. Sci Rep. 2018. PMID: 29599445 Free PMC article.
-
Yeast-based assays for characterization of the functional effects of single nucleotide polymorphisms in human DNA repair genes.PLoS One. 2018 Mar 9;13(3):e0193823. doi: 10.1371/journal.pone.0193823. eCollection 2018. PLoS One. 2018. PMID: 29522548 Free PMC article.
-
Importance of Germline and Somatic Alterations in Human MRE11, RAD50, and NBN Genes Coding for MRN Complex.Int J Mol Sci. 2023 Mar 15;24(6):5612. doi: 10.3390/ijms24065612. Int J Mol Sci. 2023. PMID: 36982687 Free PMC article. Review.
-
OsMre11 Is Required for Mitosis during Rice Growth and Development.Int J Mol Sci. 2020 Dec 26;22(1):169. doi: 10.3390/ijms22010169. Int J Mol Sci. 2020. PMID: 33375295 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
