The Par3 polarity protein is an exocyst receptor essential for mammary cell survival
- PMID: 28358000
- PMCID: PMC5379108
- DOI: 10.1038/ncomms14867
The Par3 polarity protein is an exocyst receptor essential for mammary cell survival
Abstract
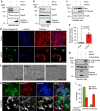
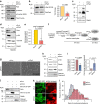
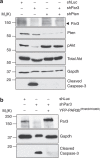
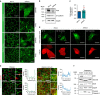
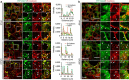
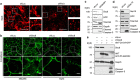
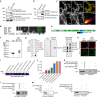
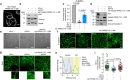
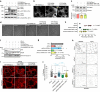
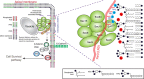
The exocyst is an essential component of the secretory pathway required for delivery of basolateral proteins to the plasma membranes of epithelial cells. Delivery occurs adjacent to tight junctions (TJ), suggesting that it recognizes a receptor at this location. However, no such receptor has been identified. The Par3 polarity protein associates with TJs but has no known function in membrane traffic. We now show that, unexpectedly, Par3 is essential for mammary cell survival. Par3 silencing causes apoptosis, triggered by phosphoinositide trisphosphate depletion and decreased Akt phosphorylation, resulting from failure of the exocyst to deliver basolateral proteins to the cortex. A small region of PAR3 binds directly to Exo70 and is sufficient for exocyst docking, membrane-protein delivery and cell survival. PAR3 lacking this domain can associate with the cortex but cannot support exocyst function. We conclude that Par3 is the long-sought exocyst receptor required for targeted membrane-protein delivery.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Shin K., Fogg V. C. & Margolis B. Tight junctions and cell polarity. Annu. Rev. Cell Dev. Biol. 22, 207–235 (2006). - PubMed
-
- Joberty G., Petersen C., Gao L. & Macara I. G. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2, 531–539 (2000). - PubMed
-
- Tabuse Y. et al.. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125, 3607–3614 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

