Phylogenomic evolutionary surveys of subtilase superfamily genes in fungi
- PMID: 28358043
- PMCID: PMC5371821
- DOI: 10.1038/srep45456
Phylogenomic evolutionary surveys of subtilase superfamily genes in fungi
Abstract
Subtilases belong to a superfamily of serine proteases which are ubiquitous in fungi and are suspected to have developed distinct functional properties to help fungi adapt to different ecological niches. In this study, we conducted a large-scale phylogenomic survey of subtilase protease genes in 83 whole genome sequenced fungal species in order to identify the evolutionary patterns and subsequent functional divergences of different subtilase families among the main lineages of the fungal kingdom. Our comparative genomic analyses of the subtilase superfamily indicated that extensive gene duplications, losses and functional diversifications have occurred in fungi, and that the four families of subtilase enzymes in fungi, including proteinase K-like, Pyrolisin, kexin and S53, have distinct evolutionary histories which may have facilitated the adaptation of fungi to a broad array of life strategies. Our study provides new insights into the evolution of the subtilase superfamily in fungi and expands our understanding of the evolution of fungi with different lifestyles.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

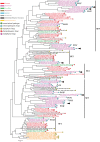

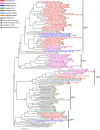
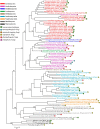
References
-
- Gunkel F. A. & Gassen H. G. Proteinase K from Tritirachium album Limber. Characterization of the chromosomal gene and expression of the cDNA in Escherichia coli. Euro J Biochem 179, 185–194 (1989). - PubMed
-
- Hu G. & St Leger R. A phylogenomic approach to reconstructing the diversification of serine proteases in fungi. J Evol Biol 17, 1204–1214 (2004). - PubMed
-
- Huang X., Zhao N. & Zhang K. Extracellular enzymes serving as virulence factors in nematophagous fungi involved in infection of the host. Res Microbiol 155, 811–816 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

