Screening for AMPA receptor auxiliary subunit specific modulators
- PMID: 28358902
- PMCID: PMC5373622
- DOI: 10.1371/journal.pone.0174742
Screening for AMPA receptor auxiliary subunit specific modulators
Abstract
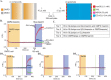
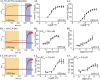
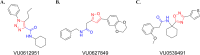
AMPA receptors (AMPAR) are ligand gated ion channels critical for synaptic transmission and plasticity. Their dysfunction is implicated in a variety of psychiatric and neurological diseases ranging from major depressive disorder to amyotrophic lateral sclerosis. Attempting to potentiate or depress AMPAR activity is an inherently difficult balancing act between effective treatments and debilitating side effects. A newly explored strategy to target subsets of AMPARs in the central nervous system is to identify compounds that affect specific AMPAR-auxiliary subunit complexes. This exploits diverse spatio-temporal expression patterns of known AMPAR auxiliary subunits, providing means for designing brain region-selective compounds. Here we report a high-throughput screening-based pipeline that can identify compounds that are selective for GluA2-CNIH3 and GluA2-stargazin complexes. These compounds will help us build upon the growing library of AMPAR-auxiliary subunit specific inhibitors, which have thus far all been targeted to TARP γ-8. We used a cell-based assay combined with a voltage-sensitive dye (VSD) to identify changes in glutamate-gated cation flow across the membranes of HEK cells co-expressing GluA2 and an auxiliary subunit. We then used a calcium flux assay to further validate hits picked from the VSD assay. VU0612951 and VU0627849 are candidate compounds from the initial screen that were identified as negative and positive allosteric modulators (NAM and PAM), respectively. They both have lower IC50/EC50s on complexes containing stargazin and CNIH3 than GSG1L or the AMPAR alone. We have also identified a candidate compound, VU0539491, that has NAM activity in GluA2(R)-CNIH3 and GluA2(Q) complexes and PAM activity in GluA2(Q)-GSG1L complexes.
Conflict of interest statement
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

