Enhancement of Arabidopsis growth characteristics using genome interrogation with artificial transcription factors
- PMID: 28358915
- PMCID: PMC5373528
- DOI: 10.1371/journal.pone.0174236
Enhancement of Arabidopsis growth characteristics using genome interrogation with artificial transcription factors
Abstract
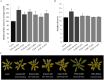
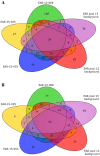
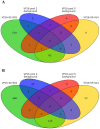
The rapidly growing world population has a greatly increasing demand for plant biomass, thus creating a great interest in the development of methods to enhance the growth and biomass accumulation of crop species. In this study, we used zinc finger artificial transcription factor (ZF-ATF)-mediated genome interrogation to manipulate the growth characteristics and biomass of Arabidopsis plants. We describe the construction of two collections of Arabidopsis lines expressing fusions of three zinc fingers (3F) to the transcriptional repressor motif EAR (3F-EAR) or the transcriptional activator VP16 (3F-VP16), and the characterization of their growth characteristics. In total, six different 3F-ATF lines with a consistent increase in rosette surface area (RSA) of up to 55% were isolated. For two lines we demonstrated that 3F-ATF constructs function as dominant in trans acting causative agents for an increase in RSA and biomass, and for five larger plant lines we have investigated 3F-ATF induced transcriptomic changes. Our results indicate that genome interrogation can be used as a powerful tool for the manipulation of plant growth and biomass and that it might supply novel cues for the discovery of genes and pathways involved in these properties.
Conflict of interest statement
Figures




References
-
- Wiebe K. The State of Food and Agriculture 2008 Biofuels: Prospects, Risks and Opportunities. Food and Agriculture Organization of the United Nations, Rome, Italy: 2008.
-
- Peng SB, Tang QY, Zou YB. Current Status and Challenges of Rice Production in China. Plant Prod Sci. 2009;12(1):3–8.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

