Bromodomain protein 4 discriminates tissue-specific super-enhancers containing disease-specific susceptibility loci in prostate and breast cancer
- PMID: 28359301
- PMCID: PMC5374680
- DOI: 10.1186/s12864-017-3620-y
Bromodomain protein 4 discriminates tissue-specific super-enhancers containing disease-specific susceptibility loci in prostate and breast cancer
Abstract
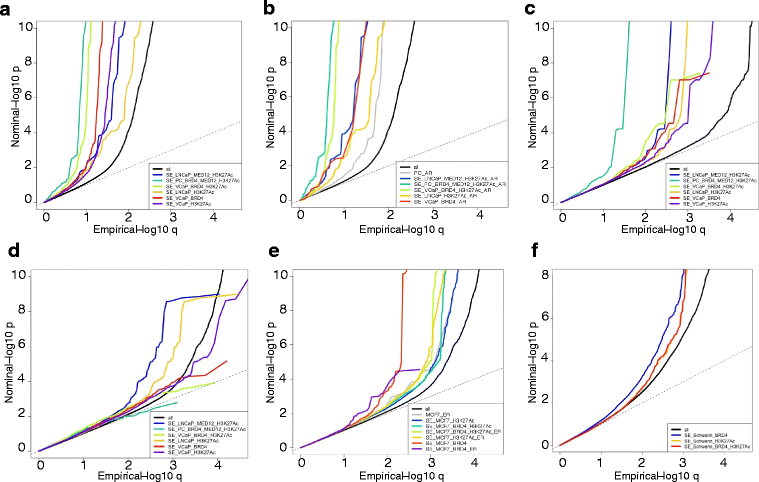
Background: Epigenetic information can be used to identify clinically relevant genomic variants single nucleotide polymorphisms (SNPs) of functional importance in cancer development. Super-enhancers are cell-specific DNA elements, acting to determine tissue or cell identity and driving tumor progression. Although previous approaches have been tried to explain risk associated with SNPs in regulatory DNA elements, so far epigenetic readers such as bromodomain containing protein 4 (BRD4) and super-enhancers have not been used to annotate SNPs. In prostate cancer (PC), androgen receptor (AR) binding sites to chromatin have been used to inform functional annotations of SNPs.
Results: Here we establish criteria for enhancer mapping which are applicable to other diseases and traits to achieve the optimal tissue-specific enrichment of PC risk SNPs. We used stratified Q-Q plots and Fisher test to assess the differential enrichment of SNPs mapping to specific categories of enhancers. We find that BRD4 is the key discriminant of tissue-specific enhancers, showing that it is more powerful than AR binding information to capture PC specific risk loci, and can be used with similar effect in breast cancer (BC) and applied to other diseases such as schizophrenia.
Conclusions: This is the first study to evaluate the enrichment of epigenetic readers in genome-wide associations studies for SNPs within enhancers, and provides a powerful tool for enriching and prioritizing PC and BC genetic risk loci. Our study represents a proof of principle applicable to other diseases and traits that can be used to redefine molecular mechanisms of human phenotypic variation.
Keywords: BRD4; Chromatin; Functional annotation; Genome-wide association studies; Prostate cancer risk; Risk loci; SNPs; breast cancer risk; schizophrenia; super-enhancer.
Figures



References
MeSH terms
Substances
Grants and funding
- G0500966/MRC_/Medical Research Council/United Kingdom
- C522/A8649/CRUK_/Cancer Research UK/United Kingdom
- R01 AG031224/AG/NIA NIH HHS/United States
- C1287/A10710/CRUK_/Cancer Research UK/United Kingdom
- MR/N003284/1/MRC_/Medical Research Council/United Kingdom
- 16565/CRUK_/Cancer Research UK/United Kingdom
- R01 EB000790/EB/NIBIB NIH HHS/United States
- G1000143/MRC_/Medical Research Council/United Kingdom
- C5047/A10692/CRUK_/Cancer Research UK/United Kingdom
- C5047/A3354/CRUK_/Cancer Research UK/United Kingdom
- R01 CA128978/CA/NCI NIH HHS/United States
- G0401527/MRC_/Medical Research Council/United Kingdom
- C5047/A15007/CRUK_/Cancer Research UK/United Kingdom
- U19 CA148065/CA/NCI NIH HHS/United States
- C1281/A12014/CRUK_/Cancer Research UK/United Kingdom
- C8197/A16565/CRUK_/Cancer Research UK/United Kingdom
- C1287/A10118/CRUK_/Cancer Research UK/United Kingdom
- 12292/A11174/CRUK_/Cancer Research UK/United Kingdom
- C5047/A8384/CRUK_/Cancer Research UK/United Kingdom
- 14136/CRUK_/Cancer Research UK/United Kingdom
- U19 CA148537/CA/NCI NIH HHS/United States
- 19170/CRUK_/Cancer Research UK/United Kingdom
- U19 CA148112/CA/NCI NIH HHS/United States
- 16561/CRUK_/Cancer Research UK/United Kingdom
- 10124/CRUK_/Cancer Research UK/United Kingdom
- RC2 DA029475/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

