Chromosomal integration of HHV-6A during non-productive viral infection
- PMID: 28360414
- PMCID: PMC5428774
- DOI: 10.1038/s41598-017-00658-y
Chromosomal integration of HHV-6A during non-productive viral infection
Abstract
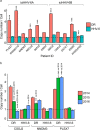
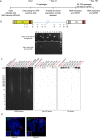
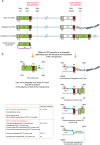
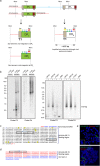
Human herpesvirus 6A (HHV-6A) and 6B (HHV-6B) are two different species of betaherpesviruses that integrate into sub-telomeric ends of human chromosomes, for which different prevalence rates of integration have been reported. It has been demonstrated that integrated viral genome is stable and is fully retained. However, study of chromosomally integrated viral genome in individuals carrying inherited HHV-6 (iciHHV-6) showed unexpected number of viral DR copies. Hence, we created an in vitro infection model and studied retention of full or partial viral genome over a period of time. We observed an exceptional event where cells retained viral direct repeats (DRs) alone in the absence of the full viral genome. Finally, we found evidence for non-telomeric integration of HHV-6A DR in both cultured cells and in an iciHHV-6 individual. Our results shed light on several novel features of HHV-6A chromosomal integration and provide valuable information for future screening techniques.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Chromosomal Integration by Human Herpesviruses 6A and 6B.Adv Exp Med Biol. 2018;1045:209-226. doi: 10.1007/978-981-10-7230-7_10. Adv Exp Med Biol. 2018. PMID: 29896669 Review.
-
The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro.Proc Natl Acad Sci U S A. 2010 Mar 23;107(12):5563-8. doi: 10.1073/pnas.0913586107. Epub 2010 Mar 8. Proc Natl Acad Sci U S A. 2010. PMID: 20212114 Free PMC article.
-
Inherited Chromosomally Integrated Human Herpesvirus 6 Demonstrates Tissue-Specific RNA Expression In Vivo That Correlates with an Increased Antibody Immune Response.J Virol. 2019 Dec 12;94(1):e01418-19. doi: 10.1128/JVI.01418-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597766 Free PMC article.
-
Latency, Integration, and Reactivation of Human Herpesvirus-6.Viruses. 2017 Jul 24;9(7):194. doi: 10.3390/v9070194. Viruses. 2017. PMID: 28737715 Free PMC article. Review.
-
Complete Genome Sequence of Germline Chromosomally Integrated Human Herpesvirus 6A and Analyses Integration Sites Define a New Human Endogenous Virus with Potential to Reactivate as an Emerging Infection.Viruses. 2016 Jan 15;8(1):19. doi: 10.3390/v8010019. Viruses. 2016. PMID: 26784220 Free PMC article.
Cited by
-
HHV-6 encoded small non-coding RNAs define an intermediate and early stage in viral reactivation.NPJ Genom Med. 2018 Sep 5;3:25. doi: 10.1038/s41525-018-0064-5. eCollection 2018. NPJ Genom Med. 2018. PMID: 30210807 Free PMC article.
-
The U94 Gene of Human Herpesvirus 6: A Narrative Review of Its Role and Potential Functions.Cells. 2020 Dec 4;9(12):2608. doi: 10.3390/cells9122608. Cells. 2020. PMID: 33291793 Free PMC article. Review.
-
Endogenization and excision of human herpesvirus 6 in human genomes.PLoS Genet. 2020 Aug 10;16(8):e1008915. doi: 10.1371/journal.pgen.1008915. eCollection 2020 Aug. PLoS Genet. 2020. PMID: 32776928 Free PMC article.
-
Chromosomally Integrated Human Herpesvirus 6: Models of Viral Genome Release from the Telomere and Impacts on Human Health.Viruses. 2017 Jul 12;9(7):184. doi: 10.3390/v9070184. Viruses. 2017. PMID: 28704957 Free PMC article. Review.
-
Chlamydia trachomatis and human herpesvirus 6 infections in ovarian cancer-Casual or causal?PLoS Pathog. 2019 Nov 7;15(11):e1008055. doi: 10.1371/journal.ppat.1008055. eCollection 2019 Nov. PLoS Pathog. 2019. PMID: 31697787 Free PMC article. Review. No abstract available.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

