Jmjd6, a JmjC Dioxygenase with Many Interaction Partners and Pleiotropic Functions
- PMID: 28360925
- PMCID: PMC5352680
- DOI: 10.3389/fgene.2017.00032
Jmjd6, a JmjC Dioxygenase with Many Interaction Partners and Pleiotropic Functions
Abstract
Lysyl hydroxylation and arginyl demethylation are post-translational events that are important for many cellular processes. The jumonji domain containing protein 6 (JMJD6) has been reported to catalyze both lysyl hydroxylation and arginyl demethylation on diverse protein substrates. It also interacts directly with RNA. This review summarizes knowledge of JMJD6 functions that have emerged in the last 15 years and considers how a single Jumonji C (JmjC) domain-containing enzyme can target so many different substrates. New links and synergies between the three main proposed functions of Jmjd6 in histone demethylation, promoter proximal pause release of polymerase II and RNA splicing are discussed. The physiological context of the described molecular functions is considered and recently described novel roles for JMJD6 in cancer and immune biology are reviewed. The increased knowledge of JMJD6 functions has wider implications for our general understanding of the JmjC protein family of which JMJD6 is a member.
Keywords: JmjC domain; PSR; chromatin; demethylation; hydroxylation; pausing; splicing; transcription.
Figures

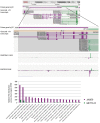


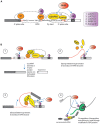
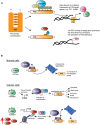
References
-
- Aprelikova O., Chen K., El Touny L. H., Brignatz-Guittard C., Han J., Qiu T., et al. (2016). The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis. Clin. Epigenetics 8:38 10.1186/s13148-016-0205-6 - DOI - PMC - PubMed
-
- Barman-Aksozen J., Beguin C., Dogar A. M., Schneider-Yin X., Minder E. I. (2013). Iron availability modulates aberrant splicing of ferrochelatase through the iron- and 2-oxoglutarate dependent dioxygenase Jmjd6 and U2AF(65.). Blood Cells Mol. Dis. 51 151–161. 10.1016/j.bcmd.2013.05.008 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

