Single-nucleotide substitution T to A in the polypyrimidine stretch at the splice acceptor site of intron 9 causes exon 10 skipping in the ACAT1 gene
- PMID: 28361105
- PMCID: PMC5370231
- DOI: 10.1002/mgg3.275
Single-nucleotide substitution T to A in the polypyrimidine stretch at the splice acceptor site of intron 9 causes exon 10 skipping in the ACAT1 gene
Abstract
Background: β-ketothiolase (T2, gene symbol ACAT1) deficiency is an autosomal recessive disorder, affecting isoleucine and ketone body metabolism. We encountered a patient (GK03) with T2 deficiency whose T2 mRNA level was <10% of the control, but in whom a previous routine cDNA analysis had failed to find any mutations. Genomic PCR-direct sequencing showed homozygosity for c.941-9T>A in the polypyrimidine stretch at the splice acceptor site of intron 9 of ACAT1. Initially, we regarded this variant as not being disease-causing by a method of predicting the effect of splicing using in silico tools. However, based on other findings of exon 10 splicing, we eventually hypothesized that this mutation causes exon 10 skipping.
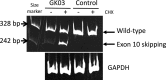
Methods: cDNA analysis was performed using GK03's fibroblasts treated with/without cycloheximide (CHX), since exon 10 skipping caused a frameshift and nonsense-mediated mRNA decay (NMD). Minigene splicing experiment was done to confirm aberrant splicing.
Results: cDNA analysis using fibroblasts cultured with cycloheximide indeed showed the occurrence of exon 10 skipping. A minigene splicing experiment clearly showed that the c.941-9T>A mutant resulted in transcripts with exon 10 skipping. There are few reports describing that single-nucleotide substitutions in polypyrimidine stretches of splice acceptor sites cause aberrant splicing.
Conclusion: We showed that c.941-9T>A induces aberrant splicing in the ACAT1 gene. Our ability to predict the effects of mutations on splicing using in silico tools is still limited. cDNA analysis and minigene splicing experiments remain useful alternatives to reveal splice defects.
Keywords: ACAT1; T2 deficiency; exon skipping; mitochondrial acetoacetyl‐CoA thiolase deficiency; polypyrimidine stretch; splice acceptor site.
Figures


Similar articles
-
Mutation update on ACAT1 variants associated with mitochondrial acetoacetyl-CoA thiolase (T2) deficiency.Hum Mutat. 2019 Oct;40(10):1641-1663. doi: 10.1002/humu.23831. Epub 2019 Jul 3. Hum Mutat. 2019. PMID: 31268215 Free PMC article. Review.
-
A novel mutation (c.121‑13T>A) in the polypyrimidine tract of the splice acceptor site of intron 2 causes exon 3 skipping in mitochondrial acetoacetyl-CoA thiolase gene.Mol Med Rep. 2017 Jun;15(6):3879-3884. doi: 10.3892/mmr.2017.6434. Epub 2017 Apr 4. Mol Med Rep. 2017. PMID: 28393214
-
Exon 10 skipping in ACAT1 caused by a novel c.949G>A mutation located at an exonic splice enhancer site.Mol Med Rep. 2016 Nov;14(5):4906-4910. doi: 10.3892/mmr.2016.5819. Epub 2016 Oct 10. Mol Med Rep. 2016. PMID: 27748876
-
A novel mutation (c.951C>T) in an exonic splicing enhancer results in exon 10 skipping in the human mitochondrial acetoacetyl-CoA thiolase gene.Mol Genet Metab. 2010 Aug;100(4):339-44. doi: 10.1016/j.ymgme.2010.03.012. Epub 2010 Mar 19. Mol Genet Metab. 2010. PMID: 20488739
-
Splicing abnormalities in congenital myasthenic syndromes.Acta Myol. 2005 Oct;24(2):50-4. Acta Myol. 2005. PMID: 16550914 Review.
Cited by
-
SLC4A4 compound heterozygous mutations in exon-intron boundary regions presenting with severe proximal renal tubular acidosis and extrarenal symptoms coexisting with Turner's syndrome: a case report.BMC Med Genet. 2018 Jun 18;19(1):103. doi: 10.1186/s12881-018-0612-y. BMC Med Genet. 2018. PMID: 29914390 Free PMC article.
-
Mutation update on ACAT1 variants associated with mitochondrial acetoacetyl-CoA thiolase (T2) deficiency.Hum Mutat. 2019 Oct;40(10):1641-1663. doi: 10.1002/humu.23831. Epub 2019 Jul 3. Hum Mutat. 2019. PMID: 31268215 Free PMC article. Review.
-
Clinical and Mutation Analysis of Patients with Best Vitelliform Macular Dystrophy or Autosomal Recessive Bestrophinopathy in Chinese Population.Biomed Res Int. 2018 Oct 18;2018:4582816. doi: 10.1155/2018/4582816. eCollection 2018. Biomed Res Int. 2018. PMID: 30498755 Free PMC article.
-
Recent advances in understanding beta-ketothiolase (mitochondrial acetoacetyl-CoA thiolase, T2) deficiency.J Hum Genet. 2019 Feb;64(2):99-111. doi: 10.1038/s10038-018-0524-x. Epub 2018 Nov 5. J Hum Genet. 2019. PMID: 30393371 Review.
References
-
- Ben Rhouma, F. , Azzouz H., Petit F. M., Khelifa M. B., Chehida A. B., Nasrallah F., et al. 2013. Molecular and biochemical characterization of a novel intronic single point mutation in a Tunisian family with glycogen storage disease type III. Mol. Biol. Rep. 40:4197–4202. - PubMed
-
- Carboni, N. , Floris M., Mateddu A., Porcu M., Marrosu G., Solla E., et al. 2011. Aberrant splicing in the LMNA gene caused by a novel mutation on the polypyrimidine tract of intron 5. Muscle Nerve 43:688–693. - PubMed
-
- Daum, R. S. , Lamm P. H., Mamer O. A., and Scriver C. R.. 1971. A “new” disorder of isoleucine catabolism. Lancet 2:1289–1290. - PubMed
-
- Fukao, T. , Yamaguchi S., Kano M., Orii T., Fujiki Y., Osumi T., et al. 1990. Molecular cloning and sequence of the complementary DNA encoding human mitochondrial acetoacetyl‐coenzyme A thiolase and study of the variant enzymes in cultured fibroblasts from patients with 3‐ketothiolase deficiency. J. Clin. Invest. 86:2086–2092. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

