Prior knowledge guided active modules identification: an integrated multi-objective approach
- PMID: 28361699
- PMCID: PMC5374590
- DOI: 10.1186/s12918-017-0388-2
Prior knowledge guided active modules identification: an integrated multi-objective approach
Abstract
Background: Active module, defined as an area in biological network that shows striking changes in molecular activity or phenotypic signatures, is important to reveal dynamic and process-specific information that is correlated with cellular or disease states.
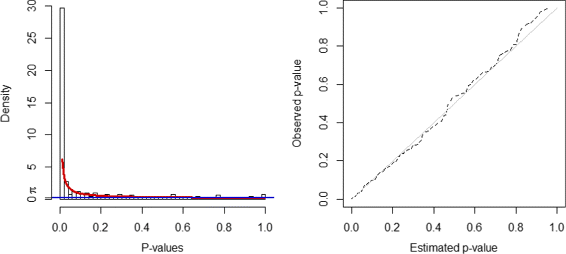
Methods: A prior information guided active module identification approach is proposed to detect modules that are both active and enriched by prior knowledge. We formulate the active module identification problem as a multi-objective optimisation problem, which consists two conflicting objective functions of maximising the coverage of known biological pathways and the activity of the active module simultaneously. Network is constructed from protein-protein interaction database. A beta-uniform-mixture model is used to estimate the distribution of p-values and generate scores for activity measurement from microarray data. A multi-objective evolutionary algorithm is used to search for Pareto optimal solutions. We also incorporate a novel constraints based on algebraic connectivity to ensure the connectedness of the identified active modules.
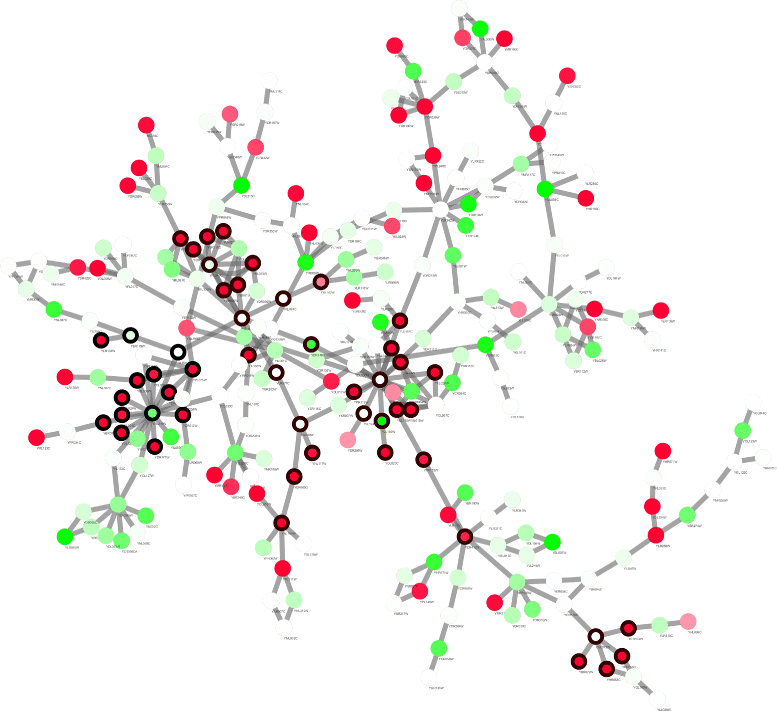
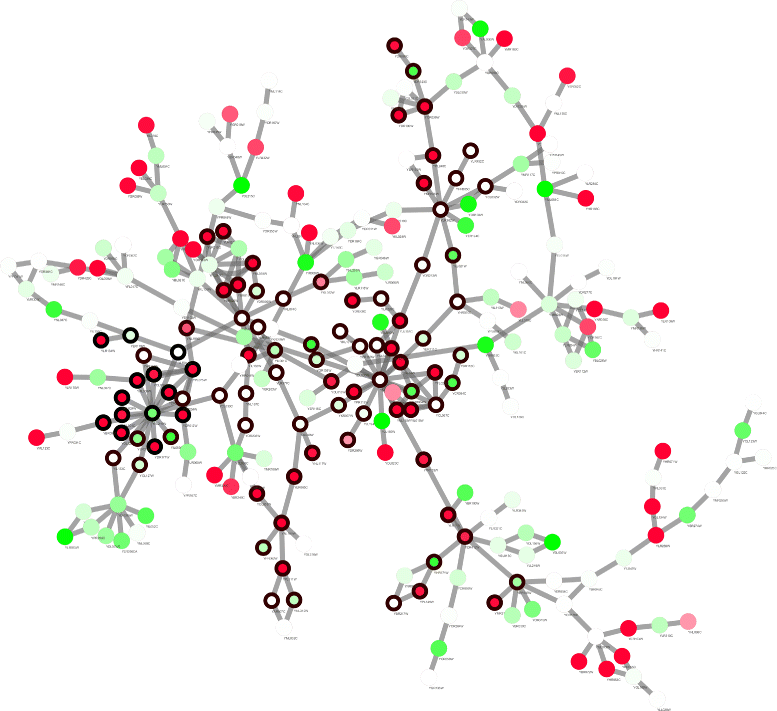
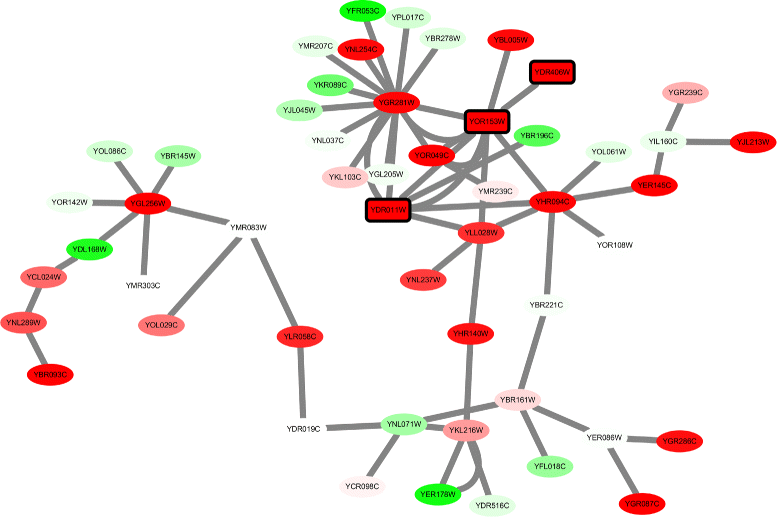
Results: Application of proposed algorithm on a small yeast molecular network shows that it can identify modules with high activities and with more cross-talk nodes between related functional groups. The Pareto solutions generated by the algorithm provides solutions with different trade-off between prior knowledge and novel information from data. The approach is then applied on microarray data from diclofenac-treated yeast cells to build network and identify modules to elucidate the molecular mechanisms of diclofenac toxicity and resistance. Gene ontology analysis is applied to the identified modules for biological interpretation.
Conclusions: Integrating knowledge of functional groups into the identification of active module is an effective method and provides a flexible control of balance between pure data-driven method and prior information guidance.
Keywords: Active module identification; Multi-objective evolutionary algorithm; Prior knowlege.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

