Genomic structure of the horse major histocompatibility complex class II region resolved using PacBio long-read sequencing technology
- PMID: 28361880
- PMCID: PMC5374520
- DOI: 10.1038/srep45518
Genomic structure of the horse major histocompatibility complex class II region resolved using PacBio long-read sequencing technology
Abstract
The mammalian Major Histocompatibility Complex (MHC) region contains several gene families characterized by highly polymorphic loci with extensive nucleotide diversity, copy number variation of paralogous genes, and long repetitive sequences. This structural complexity has made it difficult to construct a reliable reference sequence of the horse MHC region. In this study, we used long-read single molecule, real-time (SMRT) sequencing technology from Pacific Biosciences (PacBio) to sequence eight Bacterial Artificial Chromosome (BAC) clones spanning the horse MHC class II region. The final assembly resulted in a 1,165,328 bp continuous gap free sequence with 35 manually curated genomic loci of which 23 were considered to be functional and 12 to be pseudogenes. In comparison to the MHC class II region in other mammals, the corresponding region in horse shows extraordinary copy number variation and different relative location and directionality of the Eqca-DRB, -DQA, -DQB and -DOB loci. This is the first long-read sequence assembly of the horse MHC class II region with rigorous manual gene annotation, and it will serve as an important resource for association studies of immune-mediated equine diseases and for evolutionary analysis of genetic diversity in this region.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
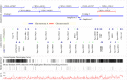



References
-
- Trowsdale J. “Both man & bird & beast”: comparative organization of MHC genes. Immunogenetics 41, 1–17 (1995). - PubMed
-
- The MHC sequencing consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature 401, 921–923, doi: http://www.nature.com/nature/journal/v401/n6756/suppinfo/401921a0_S1.html (1999). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

