Facilitating Neuron-Specific Genetic Manipulations in Drosophila melanogaster Using a Split GAL4 Repressor
- PMID: 28363977
- PMCID: PMC5499185
- DOI: 10.1534/genetics.116.199687
Facilitating Neuron-Specific Genetic Manipulations in Drosophila melanogaster Using a Split GAL4 Repressor
Abstract
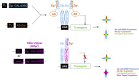
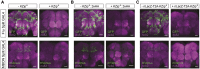
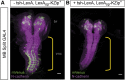
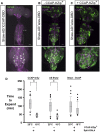
Efforts to map neural circuits have been galvanized by the development of genetic technologies that permit the manipulation of targeted sets of neurons in the brains of freely behaving animals. The success of these efforts relies on the experimenter's ability to target arbitrarily small subsets of neurons for manipulation, but such specificity of targeting cannot routinely be achieved using existing methods. In Drosophila melanogaster, a widely-used technique for refined cell type-specific manipulation is the Split GAL4 system, which augments the targeting specificity of the binary GAL4-UAS (Upstream Activating Sequence) system by making GAL4 transcriptional activity contingent upon two enhancers, rather than one. To permit more refined targeting, we introduce here the "Killer Zipper" (KZip+), a suppressor that makes Split GAL4 targeting contingent upon a third enhancer. KZip+ acts by disrupting both the formation and activity of Split GAL4 heterodimers, and we show how this added layer of control can be used to selectively remove unwanted cells from a Split GAL4 expression pattern or to subtract neurons of interest from a pattern to determine their requirement in generating a given phenotype. To facilitate application of the KZip+ technology, we have developed a versatile set of LexAop-KZip+ fly lines that can be used directly with the large number of LexA driver lines with known expression patterns. KZip+ significantly sharpens the precision of neuronal genetic control available in Drosophila and may be extended to other organisms where Split GAL4-like systems are used.
Keywords: Drosophila; Gal4-UAS; LexA-LexAop; neural circuits; transgene expression.
Copyright © 2017 Dolan et al.
Figures

Similar articles
-
Cre-assisted fine-mapping of neural circuits using orthogonal split inteins.Elife. 2020 Apr 14;9:e53041. doi: 10.7554/eLife.53041. Elife. 2020. PMID: 32286225 Free PMC article.
-
Expanding the Drosophila toolkit for dual control of gene expression.Elife. 2024 Apr 3;12:RP94073. doi: 10.7554/eLife.94073. Elife. 2024. PMID: 38569007 Free PMC article.
-
The GAL4 System: A Versatile System for the Manipulation and Analysis of Gene Expression.Methods Mol Biol. 2016;1478:33-52. doi: 10.1007/978-1-4939-6371-3_2. Methods Mol Biol. 2016. PMID: 27730574 Review.
-
A split-GAL4 driver line resource for Drosophila neuron types.Elife. 2025 Jan 24;13:RP98405. doi: 10.7554/eLife.98405. Elife. 2025. PMID: 39854223 Free PMC article.
-
The Drosophila Split Gal4 System for Neural Circuit Mapping.Front Neural Circuits. 2020 Nov 9;14:603397. doi: 10.3389/fncir.2020.603397. eCollection 2020. Front Neural Circuits. 2020. PMID: 33240047 Free PMC article. Review.
Cited by
-
Cre-assisted fine-mapping of neural circuits using orthogonal split inteins.Elife. 2020 Apr 14;9:e53041. doi: 10.7554/eLife.53041. Elife. 2020. PMID: 32286225 Free PMC article.
-
Insights from intoxicated Drosophila.Alcohol. 2019 Feb;74:21-27. doi: 10.1016/j.alcohol.2018.03.004. Epub 2018 Mar 21. Alcohol. 2019. PMID: 29980341 Free PMC article. Review.
-
A Statistically Representative Atlas for Mapping Neuronal Circuits in the Drosophila Adult Brain.Front Neuroinform. 2018 Mar 23;12:13. doi: 10.3389/fninf.2018.00013. eCollection 2018. Front Neuroinform. 2018. PMID: 29628885 Free PMC article.
-
The Role of Mitochondria in Optic Atrophy With Autosomal Inheritance.Front Neurosci. 2021 Nov 15;15:784987. doi: 10.3389/fnins.2021.784987. eCollection 2021. Front Neurosci. 2021. PMID: 34867178 Free PMC article. Review.
-
The Hunchback temporal transcription factor determines motor neuron axon and dendrite targeting in Drosophila.Development. 2019 Apr 5;146(7):dev175570. doi: 10.1242/dev.175570. Development. 2019. PMID: 30890568 Free PMC article.
References
-
- Bidaye S. S., Machacek C., Wu Y., Dickson B. J., 2014. Neuronal control of Drosophila walking direction. Science 344: 97–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

