Tissue-Specific Signaling Networks Rewired by Major Somatic Mutations in Human Cancer Revealed by Proteome-Wide Discovery
- PMID: 28364002
- PMCID: PMC5476506
- DOI: 10.1158/0008-5472.CAN-16-2460
Tissue-Specific Signaling Networks Rewired by Major Somatic Mutations in Human Cancer Revealed by Proteome-Wide Discovery
Abstract
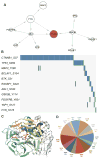
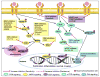
Massive somatic mutations discovered by large cancer genome sequencing projects provide unprecedented opportunities in the development of precision oncology. However, deep understanding of functional consequences of somatic mutations and identifying actionable mutations and the related drug responses currently remain formidable challenges. Dysfunction of protein posttranslational modification plays critical roles in tumorigenesis and drug responses. In this study, we proposed a novel computational oncoproteomics approach, named kinome-wide network module for cancer pharmacogenomics (KNMPx), for identifying actionable mutations that rewired signaling networks and further characterized tumorigenesis and anticancer drug responses. Specifically, we integrated 746,631 missense mutations in 4,997 tumor samples across 16 major cancer types/subtypes from The Cancer Genome Atlas into over 170,000 carefully curated nonredundant phosphorylation sites covering 18,610 proteins. We found 47 mutated proteins (e.g., ERBB2, TP53, and CTNNB1) that had enriched missense mutations at their phosphorylation sites in pan-cancer analysis. In addition, tissue-specific kinase-substrate interaction modules altered by somatic mutations identified by KNMPx were significantly associated with patient survival. We further reported a kinome-wide landscape of pharmacogenomic interactions by incorporating somatic mutation-rewired signaling networks in 1,001 cancer cell lines via KNMPx. Interestingly, we found that cell lines could highly reproduce oncogenic phosphorylation site mutations identified in primary tumors, supporting the confidence in their associations with sensitivity/resistance of inhibitors targeting EGF, MAPK, PI3K, mTOR, and Wnt signaling pathways. In summary, our KNMPx approach is powerful for identifying oncogenic alterations via rewiring phosphorylation-related signaling networks and drug sensitivity/resistance in the era of precision oncology. Cancer Res; 77(11); 2810-21. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures





Similar articles
-
Proteome-Scale Investigation of Protein Allosteric Regulation Perturbed by Somatic Mutations in 7,000 Cancer Genomes.Am J Hum Genet. 2017 Jan 5;100(1):5-20. doi: 10.1016/j.ajhg.2016.09.020. Epub 2016 Dec 8. Am J Hum Genet. 2017. PMID: 27939638 Free PMC article.
-
Individualized genetic network analysis reveals new therapeutic vulnerabilities in 6,700 cancer genomes.PLoS Comput Biol. 2020 Feb 26;16(2):e1007701. doi: 10.1371/journal.pcbi.1007701. eCollection 2020 Feb. PLoS Comput Biol. 2020. PMID: 32101536 Free PMC article.
-
An Atlas of the Human Kinome Reveals the Mutational Landscape Underlying Dysregulated Phosphorylation Cascades in Cancer.Cancer Res. 2016 Apr 1;76(7):1733-45. doi: 10.1158/0008-5472.CAN-15-2325-T. Epub 2016 Feb 26. Cancer Res. 2016. PMID: 26921330 Free PMC article.
-
Individualized network-based drug repositioning infrastructure for precision oncology in the panomics era.Brief Bioinform. 2017 Jul 1;18(4):682-697. doi: 10.1093/bib/bbw051. Brief Bioinform. 2017. PMID: 27296652 Review.
-
Integrated proteo-genomic approach for early diagnosis and prognosis of cancer.Cancer Lett. 2015 Dec 1;369(1):28-36. doi: 10.1016/j.canlet.2015.08.003. Epub 2015 Aug 11. Cancer Lett. 2015. PMID: 26276717 Review.
Cited by
-
Exome sequencing link mutation in RGPD4 with systemic sclerosis-associated interstitial lung disease and the low level of testosterone-an exploration study.Front Oncol. 2022 Sep 8;12:956552. doi: 10.3389/fonc.2022.956552. eCollection 2022. Front Oncol. 2022. PMID: 36158696 Free PMC article.
-
Large-scale pathogenicity prediction analysis of cancer-associated kinase mutations reveals variability in sensitivity and specificity of computational methods.Cancer Med. 2023 Aug;12(16):17468-17474. doi: 10.1002/cam4.6324. Epub 2023 Jul 6. Cancer Med. 2023. PMID: 37409618 Free PMC article.
-
Edgetic perturbation signatures represent known and novel cancer biomarkers.Sci Rep. 2020 Mar 9;10(1):4350. doi: 10.1038/s41598-020-61422-3. Sci Rep. 2020. PMID: 32152446 Free PMC article.
-
My personal mutanome: a computational genomic medicine platform for searching network perturbing alleles linking genotype to phenotype.Genome Biol. 2021 Jan 29;22(1):53. doi: 10.1186/s13059-021-02269-3. Genome Biol. 2021. PMID: 33514395 Free PMC article.
-
PertInInt: An Integrative, Analytical Approach to Rapidly Uncover Cancer Driver Genes with Perturbed Interactions and Functionalities.Cell Syst. 2020 Jul 22;11(1):63-74.e7. doi: 10.1016/j.cels.2020.06.005. Epub 2020 Jul 14. Cell Syst. 2020. PMID: 32711844 Free PMC article.
References
-
- Pawson T, Warner N. Oncogenic re-wiring of cellular signaling pathways. Oncogene. 2007;26:1268–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

