How Changes in Anti-SD Sequences Would Affect SD Sequences in Escherichia coli and Bacillus subtilis
- PMID: 28364038
- PMCID: PMC5427494
- DOI: 10.1534/g3.117.039305
How Changes in Anti-SD Sequences Would Affect SD Sequences in Escherichia coli and Bacillus subtilis
Abstract
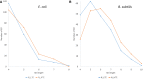
The 3' end of the small ribosomal RNAs (ssu rRNA) in bacteria is directly involved in the selection and binding of mRNA transcripts during translation initiation via well-documented interactions between a Shine-Dalgarno (SD) sequence located upstream of the initiation codon and an anti-SD (aSD) sequence at the 3' end of the ssu rRNA. Consequently, the 3' end of ssu rRNA (3'TAIL) is strongly conserved among bacterial species because a change in the region may impact the translation of many protein-coding genes. Escherichia coli and Bacillus subtilis differ in their 3' ends of ssu rRNA, being GAUCACCUCCUUA3' in E. coli and GAUCACCUCCUUUCU3' or GAUCACCUCCUUUCUA3' in B. subtilis Such differences in 3'TAIL lead to species-specific SDs (designated SDEc for E. coli and SDBs for B. subtilis) that can form strong and well-positioned SD/aSD pairing in one species but not in the other. Selection mediated by the species-specific 3'TAIL is expected to favor SDBs against SDEc in B. subtilis, but favor SDEc against SDBs in E. coli Among well-positioned SDs, SDEc is used more in E. coli than in B. subtilis, and SDBs more in B. subtilis than in E. coli Highly expressed genes and genes of high translation efficiency tend to have longer SDs than lowly expressed genes and genes with low translation efficiency in both species, but more so in B. subtilis than in E. coli Both species overuse SDs matching the bolded part of the 3'TAIL shown above. The 3'TAIL difference contributes to the host specificity of phages.
Keywords: Bacillus subtilis; Escherichia coli; Shine-Dalgarno; anti-SD-sequence; ssu rRNA; translation efficiency.
Copyright © 2017 Abolbaghaei et al.
Figures

Similar articles
-
Elucidating the 16S rRNA 3' boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data.Sci Rep. 2017 Dec 15;7(1):17639. doi: 10.1038/s41598-017-17918-6. Sci Rep. 2017. PMID: 29247194 Free PMC article.
-
The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis.Mol Microbiol. 1992 Nov;6(22):3375-83. doi: 10.1111/j.1365-2958.1992.tb02205.x. Mol Microbiol. 1992. PMID: 1283001
-
Analysis of base-pairing potentials between 16S rRNA and 5' UTR for translation initiation in various prokaryotes.Bioinformatics. 1999 Jul-Aug;15(7-8):578-81. doi: 10.1093/bioinformatics/15.7.578. Bioinformatics. 1999. PMID: 10487865
-
Regulatory RNAs in Bacillus subtilis: a Gram-Positive Perspective on Bacterial RNA-Mediated Regulation of Gene Expression.Microbiol Mol Biol Rev. 2016 Oct 26;80(4):1029-1057. doi: 10.1128/MMBR.00026-16. Print 2016 Dec. Microbiol Mol Biol Rev. 2016. PMID: 27784798 Free PMC article. Review.
-
The diversity of Shine-Dalgarno sequences sheds light on the evolution of translation initiation.RNA Biol. 2021 Nov;18(11):1489-1500. doi: 10.1080/15476286.2020.1861406. Epub 2020 Dec 21. RNA Biol. 2021. PMID: 33349119 Free PMC article. Review.
Cited by
-
Control of Translation at the Initiation Phase During Glucose Starvation in Yeast.Int J Mol Sci. 2019 Aug 19;20(16):4043. doi: 10.3390/ijms20164043. Int J Mol Sci. 2019. PMID: 31430885 Free PMC article. Review.
-
Elucidating the 16S rRNA 3' boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data.Sci Rep. 2017 Dec 15;7(1):17639. doi: 10.1038/s41598-017-17918-6. Sci Rep. 2017. PMID: 29247194 Free PMC article.
-
Unraveling the plasticity of translation initiation in prokaryotes: Beyond the invariant Shine-Dalgarno sequence.PLoS One. 2024 Jan 11;19(1):e0289914. doi: 10.1371/journal.pone.0289914. eCollection 2024. PLoS One. 2024. PMID: 38206950 Free PMC article.
-
RNA-Seq-Based Analysis Reveals Heterogeneity in Mature 16S rRNA 3' Termini and Extended Anti-Shine-Dalgarno Motifs in Bacterial Species.G3 (Bethesda). 2018 Dec 10;8(12):3973-3979. doi: 10.1534/g3.118.200729. G3 (Bethesda). 2018. PMID: 30355764 Free PMC article.
-
ComEB protein is dispensable for the transformation but must be translated for the optimal synthesis of comEC.Mol Microbiol. 2021 Jul;116(1):71-79. doi: 10.1111/mmi.14690. Epub 2021 Feb 8. Mol Microbiol. 2021. PMID: 33527432 Free PMC article.
References
-
- Agresti A., 2002. Categorical Data Analysis. Wiley, New Jersey.
-
- Bamford D. H., Caldentey J., Bamford J. K., 1995. Bacteriophage PRD1: a broad host range DSDNA tectivirus with an internal membrane. Adv. Virus Res. 45: 281–319. - PubMed
-
- Band L., Henner D. J., 1984. Bacillus subtilis requires a “stringent” Shine-Dalgarno region for gene expression. DNA 3: 17–21. - PubMed
-
- Britton R. A., Wen T., Schaefer L., Pellegrini O., Uicker W. C., et al. , 2007. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 63: 127–138. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
