Kinetic-based trapping by intervening sequence variants of the active sites of protein-disulfide isomerase identifies platelet protein substrates
- PMID: 28364042
- PMCID: PMC5454092
- DOI: 10.1074/jbc.M116.771832
Kinetic-based trapping by intervening sequence variants of the active sites of protein-disulfide isomerase identifies platelet protein substrates
Abstract
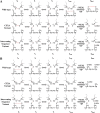
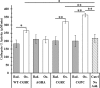
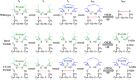
Thiol isomerases such as protein-disulfide isomerase (PDI) direct disulfide rearrangements required for proper folding of nascent proteins synthesized in the endoplasmic reticulum. Identifying PDI substrates is challenging because PDI catalyzes conformational changes that cannot be easily monitored (e.g. compared with proteolytic cleavage or amino acid phosphorylation); PDI has multiple substrates; and it can catalyze either oxidation, reduction, or isomerization of substrates. Kinetic-based substrate trapping wherein the active site motif CGHC is modified to CGHA to stabilize a PDI-substrate intermediate is effective in identifying some substrates. A limitation of this approach, however, is that it captures only substrates that are reduced by PDI, whereas many substrates are oxidized by PDI. By manipulating the highly conserved -GH- residues in the CGHC active site of PDI, we created PDI variants with a slowed reaction rate toward substrates. The prolonged intermediate state allowed us to identify protein substrates that have biased affinities for either oxidation or reduction by PDI. Because extracellular PDI is critical for thrombus formation but its extracellular substrates are not known, we evaluated the ability of these bidirectional trapping PDI variants to trap proteins released from platelets and on the platelet surface. Trapped proteins were identified by mass spectroscopy. Of the trapped substrate proteins identified by mass spectroscopy, five proteins, cathepsin G, glutaredoxin-1, thioredoxin, GP1b, and fibrinogen, showed a bias for oxidation, whereas annexin V, heparanase, ERp57, kallekrein-14, serpin B6, tetranectin, and collagen VI showed a bias for reduction. These bidirectional trapping variants will enable more comprehensive identification of thiol isomerase substrates and better elucidation of their cellular functions.
Keywords: disulfide; oxidase; platelet; protein disulfide isomerase; protein-protein interaction; reductase.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
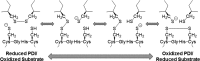
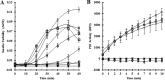
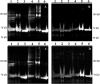
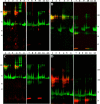
 , CGDC-PDI. RFU, relative fluorescence units.
, CGDC-PDI. RFU, relative fluorescence units.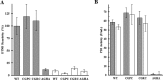


References
-
- Appenzeller-Herzog C., and Ellgaard L. (2008) The human PDI family: versatility packed into a single fold. Biochim. Biophys. Acta 1783, 535–548 - PubMed
-
- Lyles M. M., and Gilbert H. F. (1991) Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry 30, 613–619 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

