Curated protein information in the Saccharomyces genome database
- PMID: 28365727
- PMCID: PMC5467551
- DOI: 10.1093/database/bax011
Curated protein information in the Saccharomyces genome database
Abstract
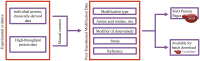
Due to recent advancements in the production of experimental proteomic data, the Saccharomyces genome database (SGD; www.yeastgenome.org ) has been expanding our protein curation activities to make new data types available to our users. Because of broad interest in post-translational modifications (PTM) and their importance to protein function and regulation, we have recently started incorporating expertly curated PTM information on individual protein pages. Here we also present the inclusion of new abundance and protein half-life data obtained from high-throughput proteome studies. These new data types have been included with the aim to facilitate cellular biology research.
Database url: : www.yeastgenome.org.
© The Author(s) 2017. Published by Oxford University Press.
Figures

References
-
- Oliveira A.P., Sauer U. (2012) The importance of post-translational modifications in regulating Saccharomyces cerevisiae metabolism. FEMS Yeast Res, 12, 104–117. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

