The 'straight mouse': defining anatomical axes in 3D embryo models
- PMID: 28365728
- PMCID: PMC5467569
- DOI: 10.1093/database/bax010
The 'straight mouse': defining anatomical axes in 3D embryo models
Abstract
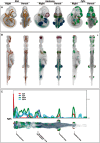
A primary objective of the eMouseAtlas Project is to enable 3D spatial mapping of whole embryo gene expression data to capture complex 3D patterns for indexing, visualization, cross-comparison and analysis. For this we have developed a spatio-temporal framework based on 3D models of embryos at different stages of development coupled with an anatomical ontology. Here we introduce a method of defining coordinate axes that correspond to the anatomical or biologically relevant anterior-posterior (A-P), dorsal-ventral (D-V) and left-right (L-R) directions. These enable more sophisticated query and analysis of the data with biologically relevant associations, and provide novel data visualizations that can reveal patterns that are otherwise difficult to detect in the standard 3D coordinate space. These anatomical coordinates are defined using the concept of a 'straight mouse-embryo' within which the anatomical coordinates are Cartesian. The straight embryo model has been mapped via a complex non-linear transform onto the standard embryo model. We explore the utility of this anatomical coordinate system in elucidating the spatial relationship of spatially mapped embryonic ' Fibroblast growth factor ' gene expression patterns, and we discuss the importance of this technology in summarizing complex multimodal mouse embryo image data from gene expression and anatomy studies.
Database url: www.emouseatlas.org.
© The Author(s) 2017. Published by Oxford University Press.
Figures


References
-
- Thompson D.W. (1917) On Growth and Form,1st edn Cambridge University Press, Cambridge.
-
- French V., Bryant P.J., Bryant S.V. (1976) Pattern regulation in epimorphic fields. Science, 193, 969–981. - PubMed
-
- Bryant S.V., French V., Bryant P.J. (1981) Distal regeneration and symmetry. Science, 212, 993–1002. - PubMed
-
- Paxinos G., Watson C. (1997) The Rat Brain in Stereotaxic Coordinates. Academic Press, San Diego, USA.
-
- Baldock R., Burger A. (2008) Anatomy ontologies: linking names to places in biology. In: Burger,A., Davidson,D., Baldock,R. (eds). Anatomy Ontologies for Bioinformatics: Principles and Practice 1st edn. Springer-Verlag London. pp. 197–212.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

