Ensembl core software resources: storage and programmatic access for DNA sequence and genome annotation
- PMID: 28365736
- PMCID: PMC5467575
- DOI: 10.1093/database/bax020
Ensembl core software resources: storage and programmatic access for DNA sequence and genome annotation
Abstract
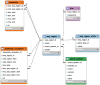
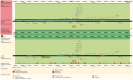
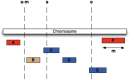
The Ensembl software resources are a stable infrastructure to store, access and manipulate genome assemblies and their functional annotations. The Ensembl 'Core' database and Application Programming Interface (API) was our first major piece of software infrastructure and remains at the centre of all of our genome resources. Since its initial design more than fifteen years ago, the number of publicly available genomic, transcriptomic and proteomic datasets has grown enormously, accelerated by continuous advances in DNA-sequencing technology. Initially intended to provide annotation for the reference human genome, we have extended our framework to support the genomes of all species as well as richer assembly models. Cross-referenced links to other informatics resources facilitate searching our database with a variety of popular identifiers such as UniProt and RefSeq. Our comprehensive and robust framework storing a large diversity of genome annotations in one location serves as a platform for other groups to generate and maintain their own tailored annotation. We welcome reuse and contributions: our databases and APIs are publicly available, all of our source code is released with a permissive Apache v2.0 licence at http://github.com/Ensembl and we have an active developer mailing list ( http://www.ensembl.org/info/about/contact/index.html ).
Database url: http://www.ensembl.org.
© The Author(s) 2017. Published by Oxford University Press.
Figures


Similar articles
-
Ensembl 2021.Nucleic Acids Res. 2021 Jan 8;49(D1):D884-D891. doi: 10.1093/nar/gkaa942. Nucleic Acids Res. 2021. PMID: 33137190 Free PMC article.
-
Ensembl 2015.Nucleic Acids Res. 2015 Jan;43(Database issue):D662-9. doi: 10.1093/nar/gku1010. Epub 2014 Oct 28. Nucleic Acids Res. 2015. PMID: 25352552 Free PMC article.
-
Using the Ensembl genome server to browse genomic sequence data.Curr Protoc Bioinformatics. 2007 Jan;Chapter 1:Unit 1.15. doi: 10.1002/0471250953.bi0115s16. Curr Protoc Bioinformatics. 2007. PMID: 18428779
-
GenomeHubs: simple containerized setup of a custom Ensembl database and web server for any species.Database (Oxford). 2017 Jan 1;2017:bax039. doi: 10.1093/database/bax039. Database (Oxford). 2017. PMID: 28605774 Free PMC article.
-
UniqTag: Content-Derived Unique and Stable Identifiers for Gene Annotation.PLoS One. 2015 May 28;10(5):e0128026. doi: 10.1371/journal.pone.0128026. eCollection 2015. PLoS One. 2015. PMID: 26020645 Free PMC article.
Cited by
-
Short and Long Noncoding RNAs Regulate the Epigenetic Status of Cells.Antioxid Redox Signal. 2018 Sep 20;29(9):832-845. doi: 10.1089/ars.2017.7262. Epub 2017 Sep 28. Antioxid Redox Signal. 2018. PMID: 28847161 Free PMC article. Review.
-
Ensembl 2020.Nucleic Acids Res. 2020 Jan 8;48(D1):D682-D688. doi: 10.1093/nar/gkz966. Nucleic Acids Res. 2020. PMID: 31691826 Free PMC article.
-
A deep ensemble framework for human essential gene prediction by integrating multi-omics data.Sci Rep. 2025 Jul 21;15(1):26407. doi: 10.1038/s41598-025-99164-9. Sci Rep. 2025. PMID: 40691502 Free PMC article.
-
RGAAT: A Reference-based Genome Assembly and Annotation Tool for New Genomes and Upgrade of Known Genomes.Genomics Proteomics Bioinformatics. 2018 Oct;16(5):373-381. doi: 10.1016/j.gpb.2018.03.006. Epub 2018 Dec 21. Genomics Proteomics Bioinformatics. 2018. PMID: 30583062 Free PMC article.
-
CADD: predicting the deleteriousness of variants throughout the human genome.Nucleic Acids Res. 2019 Jan 8;47(D1):D886-D894. doi: 10.1093/nar/gky1016. Nucleic Acids Res. 2019. PMID: 30371827 Free PMC article.
References
-
- International Human Genome Sequencing Consortium. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
Publication types
MeSH terms
Grants and funding
- WT108749/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- WT200990/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- U41 HG007823/HG/NHGRI NIH HHS/United States
- BB/M011615/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I025506/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L024225/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT201535/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- U41 HG007234/HG/NHGRI NIH HHS/United States
- B/I025360/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M01844X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 201535/Z/16/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

