Functional implications of Neandertal introgression in modern humans
- PMID: 28366169
- PMCID: PMC5376702
- DOI: 10.1186/s13059-017-1181-7
Functional implications of Neandertal introgression in modern humans
Abstract
Background: Admixture between early modern humans and Neandertals approximately 50,000-60,000 years ago has resulted in 1.5-4% Neandertal ancestry in the genomes of present-day non-Africans. Evidence is accumulating that some of these archaic alleles are advantageous for modern humans, while others are deleterious; however, the major mechanism by which these archaic alleles act has not been fully explored.
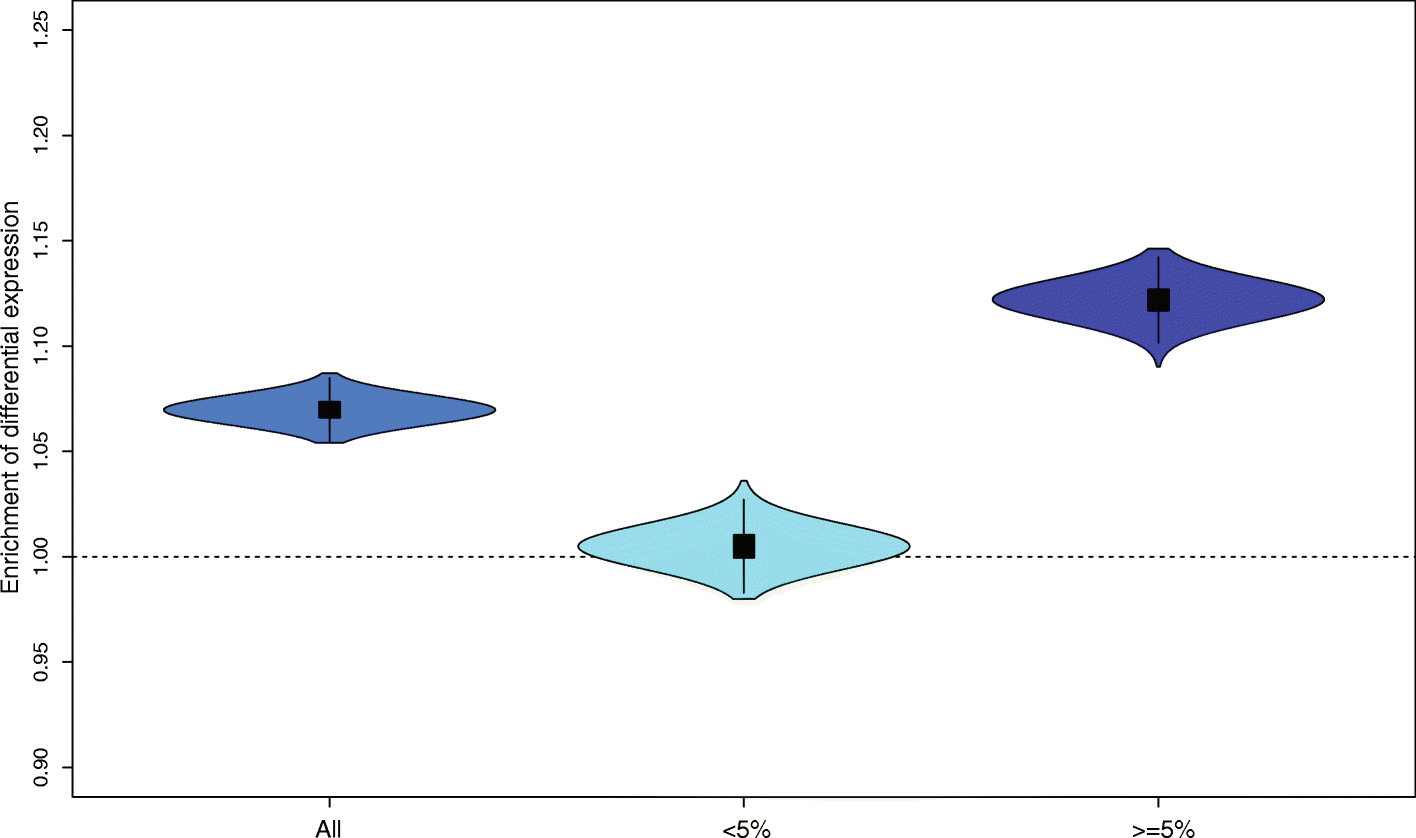
Results: Here we assess the contributions of introgressed non-synonymous and regulatory variants to modern human protein and gene expression variation. We show that gene expression changes are more often associated with Neandertal ancestry than expected, and that the introgressed non-synonymous variants tend to have less predicted functional effect on modern human proteins than mutations that arose on the human lineage. Conversely, introgressed alleles contribute proportionally more to expression variation than non-introgressed alleles.
Conclusions: Our results suggest that the major influence of Neandertal introgressed alleles is through their effects on gene regulation.
Keywords: Gene expression regulation; Human evolution; Neandertal introgression; Protein sequence variation.
Figures



Similar articles
-
Using the Neandertal genome to study the evolution of small insertions and deletions in modern humans.BMC Evol Biol. 2017 Aug 4;17(1):179. doi: 10.1186/s12862-017-1018-8. BMC Evol Biol. 2017. PMID: 28778150 Free PMC article.
-
Long-range regulatory effects of Neandertal DNA in modern humans.Genetics. 2023 Mar 2;223(3):iyac188. doi: 10.1093/genetics/iyac188. Genetics. 2023. PMID: 36560850 Free PMC article.
-
The phenotypic legacy of admixture between modern humans and Neandertals.Science. 2016 Feb 12;351(6274):737-41. doi: 10.1126/science.aad2149. Science. 2016. PMID: 26912863 Free PMC article.
-
[Progresses on Neandertal genomics].Yi Chuan. 2012 Jun;34(6):659-65. doi: 10.3724/sp.j.1005.2012.00659. Yi Chuan. 2012. PMID: 22698735 Review. Chinese.
-
The contribution of Neanderthal introgression to modern human traits.Curr Biol. 2022 Sep 26;32(18):R970-R983. doi: 10.1016/j.cub.2022.08.027. Curr Biol. 2022. PMID: 36167050 Free PMC article. Review.
Cited by
-
The genomic consequences of hybridization.Elife. 2021 Aug 4;10:e69016. doi: 10.7554/eLife.69016. Elife. 2021. PMID: 34346866 Free PMC article. Review.
-
Denisovan admixture facilitated environmental adaptation in Papua New Guinean populations.Proc Natl Acad Sci U S A. 2024 Jun 25;121(26):e2405889121. doi: 10.1073/pnas.2405889121. Epub 2024 Jun 18. Proc Natl Acad Sci U S A. 2024. PMID: 38889149 Free PMC article.
-
Archaic hominin admixture and its consequences for modern humans.Curr Opin Genet Dev. 2025 Feb;90:102280. doi: 10.1016/j.gde.2024.102280. Epub 2024 Nov 21. Curr Opin Genet Dev. 2025. PMID: 39577372 Review.
-
Biogeographic Perspectives on Human Genetic Diversification.Mol Biol Evol. 2024 Mar 1;41(3):msae029. doi: 10.1093/molbev/msae029. Mol Biol Evol. 2024. PMID: 38349332 Free PMC article.
-
A catalog of single nucleotide changes distinguishing modern humans from archaic hominins.Sci Rep. 2019 Jun 11;9(1):8463. doi: 10.1038/s41598-019-44877-x. Sci Rep. 2019. PMID: 31186485 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

