Studying the Lysine Acetylation of Malate Dehydrogenase
- PMID: 28366830
- PMCID: PMC5479488
- DOI: 10.1016/j.jmb.2017.03.027
Studying the Lysine Acetylation of Malate Dehydrogenase
Abstract
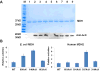
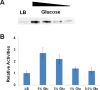
Protein acetylation plays important roles in many biological processes. Malate dehydrogenase (MDH), a key enzyme in the tricarboxylic acid cycle, has been identified to be acetylated in bacteria by proteomic studies, but no further characterization has been reported. One challenge for studying protein acetylation is to get purely acetylated proteins at specific positions. Here, we applied the genetic code expansion strategy to site-specifically incorporate Nε-acetyllysine into MDH. The acetylation of lysine residues in MDH could enhance its enzyme activity. The Escherichia coli deacetylase CobB could deacetylate acetylated MDH, while the E. coli acetyltransferase YfiQ cannot acetylate MDH efficiently. Our results also demonstrated that acetyl-CoA or acetyl-phosphate could acetylate MDH chemically in vitro. Furthermore, the acetylation level of MDH was shown to be affected by carbon sources in the growth medium.
Keywords: acetyltransferase; deacetylase; genetic code expansion; post-translational modification; protein acetylation.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures


References
-
- Cohen T, Yao TP. AcK-knowledge reversible acetylation. Sci STKE. 2004;2004:pe42. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. - PubMed
-
- Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta. 2016;1864:1372–1401. - PubMed
-
- Menzies KJ, Zhang H, Katsyuba E, Auwerx J. Protein acetylation in metabolism - metabolites and cofactors. Nat Rev Endocrinol. 2016;12:43–60. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

