Automatic single- and multi-label enzymatic function prediction by machine learning
- PMID: 28367366
- PMCID: PMC5374972
- DOI: 10.7717/peerj.3095
Automatic single- and multi-label enzymatic function prediction by machine learning
Abstract
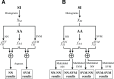
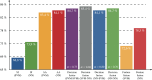
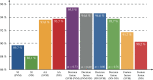
The number of protein structures in the PDB database has been increasing more than 15-fold since 1999. The creation of computational models predicting enzymatic function is of major importance since such models provide the means to better understand the behavior of newly discovered enzymes when catalyzing chemical reactions. Until now, single-label classification has been widely performed for predicting enzymatic function limiting the application to enzymes performing unique reactions and introducing errors when multi-functional enzymes are examined. Indeed, some enzymes may be performing different reactions and can hence be directly associated with multiple enzymatic functions. In the present work, we propose a multi-label enzymatic function classification scheme that combines structural and amino acid sequence information. We investigate two fusion approaches (in the feature level and decision level) and assess the methodology for general enzymatic function prediction indicated by the first digit of the enzyme commission (EC) code (six main classes) on 40,034 enzymes from the PDB database. The proposed single-label and multi-label models predict correctly the actual functional activities in 97.8% and 95.5% (based on Hamming-loss) of the cases, respectively. Also the multi-label model predicts all possible enzymatic reactions in 85.4% of the multi-labeled enzymes when the number of reactions is unknown. Code and datasets are available at https://figshare.com/s/a63e0bafa9b71fc7cbd7.
Keywords: Amino acid sequence; Enzyme classification; Multi-label; Single-label; Smith-Waterman algorithm; Structural information.
Conflict of interest statement
Nikos Paragios and Evangelia Zacharaki are employees of Equipe GALEN, INRIA Saclay, France.
Figures
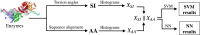

References
-
- Amidi A, Amidi S, Vlachakis D, Paragios N, Zacharaki EI. A machine learning methodology for enzyme functional classification combining structural and protein sequence descriptors. Lecture Notes in Computer Science. 2016;9656:728–738.
-
- Borgwardt KM, Ong CS, Schönauer S, Vishwanathan SVN, Smola AJ, Kriegel H-P. Protein function prediction via graph kernels. Bioinformatics. 2005;21(Suppl 1):i47–i56. - PubMed
-
- Concu R, Dea-Ayuela M, Perez-Montoto L, Bolas-Fernandez F, Prado-Prado F, Podda G, Uriarte E, Ubeira F, Gonzalez-Diaz H. Prediction of enzyme classes from 3D structure: a general model and examples of experimental-theoretic scoring of peptide mass fingerprints of leishmania proteins. Journal of Proteome Research. 2009a;8(9):4372–4382. doi: 10.1021/pr9003163. - DOI - PubMed
-
- Concu R, Dea-Ayuela M, Perez-Montoto L, Uriarte F, Bolas-Fernandez F, Podda G, Pazos A, Munteanu C, Ubeira F, Gonzalez-Diaz H. 3D entropy and moments prediction of enzyme classes and experimental-theoretic study of peptide fingerprints in leishmania parasites. Biochimica et Biophysica Acta (BBA)–Proteins and Proteomics. 2009b;1794(12):1784–1794. doi: 10.1016/j.bbapap.2009.08.020. - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

