High-resolution profiles of gene expression and DNA methylation highlight mitochondrial modifications during early embryonic development
- PMID: 28367907
- PMCID: PMC5481627
- DOI: 10.1262/jrd.2016-168
High-resolution profiles of gene expression and DNA methylation highlight mitochondrial modifications during early embryonic development
Abstract
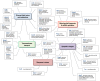
Well-organized mitochondrial functions and dynamics are critical for early embryonic development and are operated via a large number of mitochondria-related genes (MtGs) encoded by both the nuclear and the mitochondrial genome. However, the mechanisms underlying mitochondrial modifications during the critical window between blastocyst implantation and postimplantation organogenesis are poorly understood. Herein, we performed high-resolution dynamic profiling of MtGs to acquire a more detailed understanding of mitochondrial modifications during early development. Our data suggest that the resumption of mitochondrial mass growth is not only facilitated by increased mitochondrial biogenesis and mitochondrial DNA (mtDNA) replication, but also by the appropriate balance between mitochondrial fission and fusion. In addition, increased levels of reactive oxygen species (ROS) resulting from enhanced mitochondrial functions may be the critical inducer for activating the glutathione (GSH)-based stress response system in early embryos. The appropriate balance between the mitochondrial stress response and apoptosis appears to be significant for cell differentiation and early organogenesis. Furthermore, we found that most MtGs undergo de novo promoter methylation, which may have functional consequences on mitochondrial functions and dynamics during early development. We also report that mtDNA methylation can be observed as early as soon after implantation. DNMT1, the predominant enzyme for maintaining DNA methylation, localized to the mitochondria and bound to mtDNA by the implantation stage. Our study provides a new insight into the involvement of mitochondria in early mammalian embryogenesis. We also propose that the epigenetic modifications during early development are significant for modulating mitochondrial functions and dynamics.
Keywords: DNA methylation; Early embryos; Glutathione; Mitochondria; Reactive oxygen species.
Figures






References
-
- Harvey A, Gibson T, Lonergan T, Brenner C. Dynamic regulation of mitochondrial function in preimplantation embryos and embryonic stem cells. Mitochondrion 2011; 11: 829–838. - PubMed
-
- Ramalho-Santos J, Varum S, Amaral S, Mota PC, Sousa AP, Amaral A. Mitochondrial functionality in reproduction: from gonads and gametes to embryos and embryonic stem cells. Hum Reprod Update 2009; 15: 553–572. - PubMed
-
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 2003; 115: 629–640. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

