Generation of Aptamers from A Primer-Free Randomized ssDNA Library Using Magnetic-Assisted Rapid Aptamer Selection
- PMID: 28367958
- PMCID: PMC5377317
- DOI: 10.1038/srep45478
Generation of Aptamers from A Primer-Free Randomized ssDNA Library Using Magnetic-Assisted Rapid Aptamer Selection
Abstract
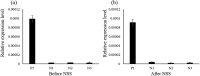
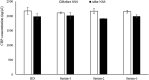
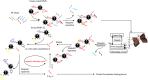
Aptamers are oligonucleotides that can bind to specific target molecules. Most aptamers are generated using random libraries in the standard systematic evolution of ligands by exponential enrichment (SELEX). Each random library contains oligonucleotides with a randomized central region and two fixed primer regions at both ends. The fixed primer regions are necessary for amplifying target-bound sequences by PCR. However, these extra-sequences may cause non-specific bindings, which potentially interfere with good binding for random sequences. The Magnetic-Assisted Rapid Aptamer Selection (MARAS) is a newly developed protocol for generating single-strand DNA aptamers. No repeat selection cycle is required in the protocol. This study proposes and demonstrates a method to isolate aptamers for C-reactive proteins (CRP) from a randomized ssDNA library containing no fixed sequences at 5' and 3' termini using the MARAS platform. Furthermore, the isolated primer-free aptamer was sequenced and binding affinity for CRP was analyzed. The specificity of the obtained aptamer was validated using blind serum samples. The result was consistent with monoclonal antibody-based nephelometry analysis, which indicated that a primer-free aptamer has high specificity toward targets. MARAS is a feasible platform for efficiently generating primer-free aptamers for clinical diagnoses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Similar articles
-
Primer-free aptamer selection using a random DNA library.J Vis Exp. 2010 Jul 26;(41):2039. doi: 10.3791/2039. J Vis Exp. 2010. PMID: 20689511 Free PMC article.
-
An improved SELEX technique for selection of DNA aptamers binding to M-type 11 of Streptococcus pyogenes.Methods. 2016 Mar 15;97:51-7. doi: 10.1016/j.ymeth.2015.12.005. Epub 2015 Dec 8. Methods. 2016. PMID: 26678795
-
[Efficient screening for 8-oxoguanine DNA glycosylase binding aptamers via capillary electrophoresis].Se Pu. 2021 Jul 8;39(7):721-729. doi: 10.3724/SP.J.1123.2020.12017. Se Pu. 2021. PMID: 34227370 Free PMC article. Chinese.
-
Inside the Black Box: What Makes SELEX Better?Molecules. 2019 Oct 7;24(19):3598. doi: 10.3390/molecules24193598. Molecules. 2019. PMID: 31591283 Free PMC article. Review.
-
The shorter the better: reducing fixed primer regions of oligonucleotide libraries for aptamer selection.Molecules. 2009 Mar 27;14(4):1353-69. doi: 10.3390/molecules14041353. Molecules. 2009. PMID: 19384268 Free PMC article. Review.
Cited by
-
Critical Design Factors for Electrochemical Aptasensors Based on Target-Induced Conformational Changes: The Case of Small-Molecule Targets.Biosensors (Basel). 2022 Oct 1;12(10):816. doi: 10.3390/bios12100816. Biosensors (Basel). 2022. PMID: 36290952 Free PMC article. Review.
-
Perspective on the Future Role of Aptamers in Analytical Chemistry.Anal Chem. 2019 Dec 17;91(24):15335-15344. doi: 10.1021/acs.analchem.9b03853. Epub 2019 Nov 26. Anal Chem. 2019. PMID: 31714748 Free PMC article.
-
Key Aspects of Nucleic Acid Library Design for in Vitro Selection.Int J Mol Sci. 2018 Feb 5;19(2):470. doi: 10.3390/ijms19020470. Int J Mol Sci. 2018. PMID: 29401748 Free PMC article. Review.
-
Simple Methods and Rational Design for Enhancing Aptamer Sensitivity and Specificity.Front Mol Biosci. 2018 May 14;5:41. doi: 10.3389/fmolb.2018.00041. eCollection 2018. Front Mol Biosci. 2018. PMID: 29868605 Free PMC article.
-
Screening and Identification of DNA Nanostructure Aptamer Using the SELEX Method for Detection of Epsilon Toxin.Iran J Pharm Res. 2023 Dec 8;22(1):e140505. doi: 10.5812/ijpr-140505. eCollection 2023 Jan-Dec. Iran J Pharm Res. 2023. PMID: 38444705 Free PMC article.
References
-
- Tuerk C. & Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 249, 505–510 (1990). - PubMed
-
- Ellington A. D. & Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 346, 818–822 (1990). - PubMed
-
- Ellington A. D. & Szostak J. W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 355, 850–852 (1992). - PubMed
-
- Mimjee S. M., Rusconi C. P. & Sullenger B. A. Aptamers: an emerging class of therapeutic. Annu Rev Med. 56, 555–583 (2005). - PubMed
-
- Hicke B. J., Stephens A. W., Gould T., Chang Y. F., Lynott C. K. et al.. Tumor targeting by an aptamer. J Nucl Med. 47, 668–678 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

