Protein-coding genes in B chromosomes of the grasshopper Eyprepocnemis plorans
- PMID: 28367986
- PMCID: PMC5377258
- DOI: 10.1038/srep45200
Protein-coding genes in B chromosomes of the grasshopper Eyprepocnemis plorans
Abstract
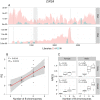
For many years, parasitic B chromosomes have been considered genetically inert elements. Here we show the presence of ten protein-coding genes in the B chromosome of the grasshopper Eyprepocnemis plorans. Four of these genes (CIP2A, GTPB6, KIF20A, and MTG1) were complete in the B chromosome whereas the six remaining (CKAP2, CAP-G, HYI, MYCB2, SLIT and TOP2A) were truncated. Five of these genes (CIP2A, CKAP2, CAP-G, KIF20A, and MYCB2) were significantly up-regulated in B-carrying individuals, as expected if they were actively transcribed from the B chromosome. This conclusion is supported by three truncated genes (CKAP2, CAP-G and MYCB2) which showed up-regulation only in the regions being present in the B chromosome. Our results indicate that B chromosomes are not so silenced as was hitherto believed. Interestingly, the five active genes in the B chromosome code for functions related with cell division, which is the main arena where B chromosome destiny is played. This suggests that B chromosome evolutionary success can lie on its gene content.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Puertas M. J. et al. Genetic control of B chromosome transmission in maize and rye. In Chromosomes Today(eds Olmo P. D. E. & Redi P. C. A.) 79–92 (Birkhäuser Basel, 2000).
-
- Camacho J. P. M. B chromosomes. In The Evolution of the Genome(ed. Gregory T. R.) 223–286 (Academic Press, 2005).
-
- Graphodatsky A. S. et al. The proto-oncogene C-KIT maps to canid B-chromosomes. Chromosome Res. 13, 113–122 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

