Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease
- PMID: 28368383
- PMCID: PMC5420447
- DOI: 10.1038/nm.4307
Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease
Erratum in
-
Erratum: Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease.Nat Med. 2017 Jun 6;23(6):788. doi: 10.1038/nm0617-788d. Nat Med. 2017. PMID: 28586341 No abstract available.
Abstract
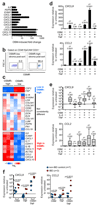
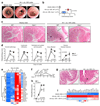
Inflammatory bowel diseases (IBD), including Crohn's disease (CD) and ulcerative colitis (UC), are complex chronic inflammatory conditions of the gastrointestinal tract that are driven by perturbed cytokine pathways. Anti-tumor necrosis factor-α (TNF) antibodies are mainstay therapies for IBD. However, up to 40% of patients are nonresponsive to anti-TNF agents, which makes the identification of alternative therapeutic targets a priority. Here we show that, relative to healthy controls, inflamed intestinal tissues from patients with IBD express high amounts of the cytokine oncostatin M (OSM) and its receptor (OSMR), which correlate closely with histopathological disease severity. The OSMR is expressed in nonhematopoietic, nonepithelial intestinal stromal cells, which respond to OSM by producing various proinflammatory molecules, including interleukin (IL)-6, the leukocyte adhesion factor ICAM1, and chemokines that attract neutrophils, monocytes, and T cells. In an animal model of anti-TNF-resistant intestinal inflammation, genetic deletion or pharmacological blockade of OSM significantly attenuates colitis. Furthermore, according to an analysis of more than 200 patients with IBD, including two cohorts from phase 3 clinical trials of infliximab and golimumab, high pretreatment expression of OSM is strongly associated with failure of anti-TNF therapy. OSM is thus a potential biomarker and therapeutic target for IBD, and has particular relevance for anti-TNF-resistant patients.
Conflict of interest statement
NRW, ANH, BMJO, and FP are inventors of patents relating to OSM as a therapeutic target for IBD. NRW, ANH, BMJO, SPLT, and FP are shareholders in ImmElpis Pharma Ltd. SK has received honoraria and/or research support from Allergan, Abbvie, Astra-Zeneca, ChemoCentryx Inc., Dr Falk Pharma, Ferring, Gilead, GSK, Merck, Mitsubishi-Tanabe Pharma, Pfizer, and Vifor Pharma. SPLT has received research support from Abbvie, IOIBD, Lilly, UCB, Vifor, and the Norman Collison Foundation. SPLT receives consulting fees from Abbvie, Amgen, Biogen, Boehringer Ingelheim, Bristol-Myers Squibb, Celgene, Chemocentryx, Cosmo, Ferring, Giuliani SpA, GSK, Lilly, MSD, Neovacs, NovoNordisk, Norman Collison Foundation, Novartis, NPS Pharmaceuticals, Pfizer, Proximagen, Receptos, Shire, Sigmoid Pharma, Takeda, Topivert, UCB, VHsquared, and Vifor Pharma. SPLT has received speaker fees from Abbvie, Biogen, Ferring, and Takeda. BL, FB, CB, and SEP are employees of Janssen Research and Development LLC. SB and MC are currently employees of GSK. FP receives consultancy fees from Janssen and GSK (unrelated to this work).
Figures




Comment in
-
IBD: Oncostatin M promotes inflammation in IBD.Nat Rev Gastroenterol Hepatol. 2017 May;14(5):261. doi: 10.1038/nrgastro.2017.47. Epub 2017 Apr 12. Nat Rev Gastroenterol Hepatol. 2017. PMID: 28400625 No abstract available.
-
Measuring inflammation in IBD with an OSM-ostat.Sci Transl Med. 2017 May 3;9(388):eaan3777. doi: 10.1126/scitranslmed.aan3777. Sci Transl Med. 2017. PMID: 28469037
-
A role for oncostatin M in inflammatory bowel disease.Nat Med. 2017 May 5;23(5):535-536. doi: 10.1038/nm.4338. Nat Med. 2017. PMID: 28475567 Free PMC article.
References
-
- Uniken Venema WT, Voskuil MD, Dijkstra G, Weersma RK, Festen EA. The genetic background of inflammatory bowel disease: from correlation to causality. J Pathol. 2017;241:146–158. - PubMed
-
- de Souza HSP, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13–27. - PubMed
-
- Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. - PubMed
-
- Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–342. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

