Protein S-acyl transferase 4 controls nucleus position during root hair tip growth
- PMID: 28368733
- PMCID: PMC5437833
- DOI: 10.1080/15592324.2017.1311438
Protein S-acyl transferase 4 controls nucleus position during root hair tip growth
Abstract
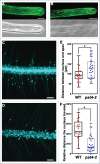
Protein S-acyl transferases (PATs) play critical roles in plant developmental and environmental responses by catalyzing S-acylation of substrate proteins, most of which are involved in cellular signaling. However, only few plant PATs have been functionally characterized. We recently demonstrated that Arabidopsis PAT4 mediates root hair elongation by positively regulating the membrane association of ROP2 and actin microfilament organization. Here, we show that apex-associated re-positioning of nucleus during root hair elongation was impaired by PAT4 loss-of-function. Results presented here pose a significant question concerning the molecular machinery mediating nuclear migration during root hair growth.
Keywords: Microtubule; nuclear positioning; palmitoylation; root hairs; tip growth.
Figures
References
-
- Greaves J, Chamberlain LH. DHHC palmitoyl transferases: Substrate interactions and (patho)physiology. Trends Biochem Sci 2011; 36:245-53; PMID:21388813; http://dx.doi.org/ 10.1016/j.tibs.2011.01.003 - DOI - PubMed
-
- Hemsley PA, Grierson CS. Multiple roles for protein palmitoylation in plants. Trends Plant Sci 2008; 13:295-302; PMID:18501662; http://dx.doi.org/ 10.1016/j.tplants.2008.04.006 - DOI - PubMed
-
- Running MP. The role of lipid post-translational modification in plant developmental processes. Front Plant Sci 2014; 5:50; PMID:24600462; http://dx.doi.org/ 10.3389/fpls.2014.00050 - DOI - PMC - PubMed
-
- Hemsley PA, Weimar T, Lilley KS, Dupree P, Grierson CS. A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol 2013; 197:805-14; PMID:23252521; http://dx.doi.org/ 10.1111/nph.12077 - DOI - PubMed
-
- Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al.. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 2008; 456:904-9; PMID:19092927; http://dx.doi.org/ 10.1038/nature07605 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
