Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival
- PMID: 28369050
- PMCID: PMC5540372
- DOI: 10.1038/ni.3716
Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival
Abstract
The transcription factor STAT5 has a critical role in B cell acute lymphoblastic leukemia (B-ALL). How STAT5 mediates this effect is unclear. Here we found that activation of STAT5 worked together with defects in signaling components of the precursor to the B cell antigen receptor (pre-BCR), including defects in BLNK, BTK, PKCβ, NF-κB1 and IKAROS, to initiate B-ALL. STAT5 antagonized the transcription factors NF-κB and IKAROS by opposing regulation of shared target genes. Super-enhancers showed enrichment for STAT5 binding and were associated with an opposing network of transcription factors, including PAX5, EBF1, PU.1, IRF4 and IKAROS. Patients with a high ratio of active STAT5 to NF-κB or IKAROS had more-aggressive disease. Our studies indicate that an imbalance of two opposing transcriptional programs drives B-ALL and suggest that restoring the balance of these pathways might inhibit B-ALL.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS: The authors have no competing financial interests.
Figures
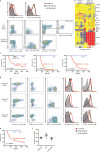
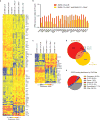
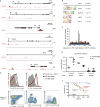
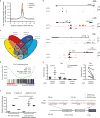



Comment in
-
It is a differentiation game: STAT5 in a new role.Cell Death Differ. 2017 Jun;24(6):953-954. doi: 10.1038/cdd.2017.71. Epub 2017 May 12. Cell Death Differ. 2017. PMID: 28498368 Free PMC article. No abstract available.
-
STAT5 alters the state of transcriptional networks, driving aggressive leukemia.Nat Immunol. 2017 May 18;18(6):597-598. doi: 10.1038/ni.3747. Nat Immunol. 2017. PMID: 28518152 No abstract available.
References
-
- Bhatia S, et al. Low incidence of second neoplasms among children diagnosed with acute lymphoblastic leukemia after 1983. Blood. 2002;99:4257–4264. - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA154998/CA/NCI NIH HHS/United States
- P50 CA100632/CA/NCI NIH HHS/United States
- R01 CA213138/CA/NCI NIH HHS/United States
- R01 CA157644/CA/NCI NIH HHS/United States
- R01 CA172558/CA/NCI NIH HHS/United States
- R01 CA169458/CA/NCI NIH HHS/United States
- R21 CA209229/CA/NCI NIH HHS/United States
- R35 CA197628/CA/NCI NIH HHS/United States
- R01 CA139032/CA/NCI NIH HHS/United States
- R01 CA185062/CA/NCI NIH HHS/United States
- R01 CA137060/CA/NCI NIH HHS/United States
- T32 AI007313/AI/NIAID NIH HHS/United States
- R01 CA151845/CA/NCI NIH HHS/United States
- P30 CA077598/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

