Origin and Evolution of the Kiwifruit Canker Pandemic
- PMID: 28369338
- PMCID: PMC5388287
- DOI: 10.1093/gbe/evx055
Origin and Evolution of the Kiwifruit Canker Pandemic
Abstract
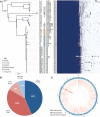
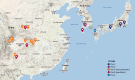
Recurring epidemics of kiwifruit (Actinidia spp.) bleeding canker disease are caused by Pseudomonas syringae pv. actinidiae (Psa). In order to strengthen understanding of population structure, phylogeography, and evolutionary dynamics, we isolated Pseudomonas from cultivated and wild kiwifruit across six provinces in China. Based on the analysis of 80 sequenced Psa genomes, we show that China is the origin of the pandemic lineage but that strain diversity in China is confined to just a single clade. In contrast, Korea and Japan harbor strains from multiple clades. Distinct independent transmission events marked introduction of the pandemic lineage into New Zealand, Chile, Europe, Korea, and Japan. Despite high similarity within the core genome and minimal impact of within-clade recombination, we observed extensive variation even within the single clade from which the global pandemic arose.
Keywords: bacterial plant pathogen; disease emergence; genomic epidemiology; pathogen evolution; plant-microbe interactions.
© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Abelleira A, et al. 2011. First report of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae in Spain. Plant Dis. 95:1583–1583. - PubMed
-
- Almeida RPP, Nunney L. 2015. How do plant diseases caused by Xylella fastidiosa emerge?. Plant Dis. 99:1457–1467. - PubMed
-
- Andam CP, Worby CJ, Chang Q, Campana MG. 2016. Microbial genomics of ancient plagues and outbreaks. Trends Microbiol. 24(12):978–990. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

