A non-catalytic role of RecBCD in homology directed gap repair and translesion synthesis
- PMID: 28369478
- PMCID: PMC5449595
- DOI: 10.1093/nar/gkx217
A non-catalytic role of RecBCD in homology directed gap repair and translesion synthesis
Abstract
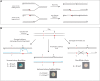
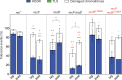
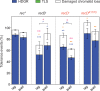
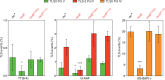
The RecBCD complex is a key factor in DNA metabolism. This protein complex harbors a processive nuclease and two helicases activities that give it the ability to process duplex DNA ends. These enzymatic activities make RecBCD a major player in double strand break repair, conjugational recombination and degradation of linear DNA. In this work, we unravel a new role of the RecBCD complex in the processing of DNA single-strand gaps that are generated at DNA replication-blocking lesions. We show that independently of its nuclease or helicase activities, the entire RecBCD complex is required for recombinational repair of the gap and efficient translesion synthesis. Since none of the catalytic functions of RecBCD are required for those processes, we surmise that the complex acts as a structural element that stabilizes the blocked replication fork, allowing efficient DNA damage tolerance.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
RecBCD is required to complete chromosomal replication: Implications for double-strand break frequencies and repair mechanisms.DNA Repair (Amst). 2015 Aug;32:86-95. doi: 10.1016/j.dnarep.2015.04.018. Epub 2015 May 2. DNA Repair (Amst). 2015. PMID: 26003632 Free PMC article. Review.
-
The hybrid recombinational repair pathway operates in a χ activity deficient recC1004 mutant of Escherichia coli.Biochimie. 2012 Sep;94(9):1918-25. doi: 10.1016/j.biochi.2012.05.008. Epub 2012 May 19. Biochimie. 2012. PMID: 22617484
-
RecBCD Enzyme "Chi Recognition" Mutants Recognize Chi Recombination Hotspots in the Right DNA Context.Genetics. 2016 Sep;204(1):139-52. doi: 10.1534/genetics.116.191056. Epub 2016 Jul 8. Genetics. 2016. PMID: 27401752 Free PMC article.
-
A model of DNA unwinding dynamics by the RecBCD complex and its regulation by Chi recognition.J Theor Biol. 2018 Jul 7;448:142-156. doi: 10.1016/j.jtbi.2018.04.014. Epub 2018 Apr 12. J Theor Biol. 2018. PMID: 29655868
-
Single-strand gap repair involves both RecF and RecBCD pathways.Curr Genet. 2016 Aug;62(3):519-21. doi: 10.1007/s00294-016-0575-5. Epub 2016 Feb 13. Curr Genet. 2016. PMID: 26874520 Review.
Cited by
-
Single strand gap repair: The presynaptic phase plays a pivotal role in modulating lesion tolerance pathways.PLoS Genet. 2022 Jun 2;18(6):e1010238. doi: 10.1371/journal.pgen.1010238. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35653392 Free PMC article.
-
DNA lesions proximity modulates damage tolerance pathways in Escherichia coli.Nucleic Acids Res. 2018 May 4;46(8):4004-4012. doi: 10.1093/nar/gky135. Nucleic Acids Res. 2018. PMID: 29529312 Free PMC article.
-
Recombination Mediator Proteins: Misnomers That Are Key to Understanding the Genomic Instabilities in Cancer.Genes (Basel). 2022 Feb 27;13(3):437. doi: 10.3390/genes13030437. Genes (Basel). 2022. PMID: 35327990 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

