Antagonistic Coevolution of MER Tyrosine Kinase Expression and Function
- PMID: 28369510
- PMCID: PMC5850725
- DOI: 10.1093/molbev/msx102
Antagonistic Coevolution of MER Tyrosine Kinase Expression and Function
Abstract
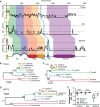
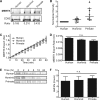
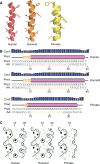
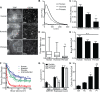
TYRO3, AXL, and MERTK (TAM) receptors are a family of receptor tyrosine kinases that maintain homeostasis through the clearance of apoptotic cells, and when defective, contribute to chronic inflammatory and autoimmune diseases such as atherosclerosis, multiple sclerosis, systemic lupus erythematosus, rheumatoid arthritis, and Crohn's disease. In addition, certain enveloped viruses utilize TAM receptors for immune evasion and entry into host cells, with several viruses preferentially hijacking MERTK for these purposes. Despite the biological importance of TAM receptors, little is understood of their recent evolution and its impact on their function. Using evolutionary analysis of primate TAM receptor sequences, we identified strong, recent positive selection in MERTK's signal peptide and transmembrane domain that was absent from TYRO3 and AXL. Reconstruction of hominid and primate ancestral MERTK sequences revealed three nonsynonymous single nucleotide polymorphisms in the human MERTK signal peptide, with a G14C mutation resulting in a predicted non-B DNA cruciform motif, producing a significant decrease in MERTK expression with no significant effect on MERTK trafficking or half-life. Reconstruction of MERTK's transmembrane domain identified three amino acid substitutions and four amino acid insertions in humans, which led to significantly higher levels of self-clustering through the creation of a new interaction motif. This clustering counteracted the effect of the signal peptide mutations through enhancing MERTK avidity, whereas the lower MERTK expression led to reduced binding of Ebola virus-like particles. The decreased MERTK expression counterbalanced by increased avidity is consistent with antagonistic coevolution to evade viral hijacking of MERTK.
Keywords: MERTK; TAM receptors; antagonistic coevolution; efferocytosis; positive selection; viral infection.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Bansal M, Kumar A, Yella VR.. 2014. Role of DNA sequence based structural features of promoters in transcription initiation and gene expression. Curr Opin Struct Biol. 25:77–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

