Non-coding RNA may be associated with cytoplasmic male sterility in Silene vulgaris
- PMID: 28369520
- PMCID: PMC5444436
- DOI: 10.1093/jxb/erx057
Non-coding RNA may be associated with cytoplasmic male sterility in Silene vulgaris
Abstract
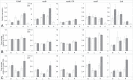
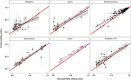
Cytoplasmic male sterility (CMS) is a widespread phenomenon in flowering plants caused by mitochondrial (mt) genes. CMS genes typically encode novel proteins that interfere with mt functions and can be silenced by nuclear fertility-restorer genes. Although the molecular basis of CMS is well established in a number of crop systems, our understanding of it in natural populations is far more limited. To identify CMS genes in a gynodioecious plant, Silene vulgaris, we constructed mt transcriptomes and compared transcript levels and RNA editing patterns in floral bud tissue from female and hermaphrodite full siblings. The transcriptomes from female and hermaphrodite individuals were very similar overall with respect to variation in levels of transcript abundance across the genome, the extent of RNA editing, and the order in which RNA editing and intron splicing events occurred. We found only a single genomic region that was highly overexpressed and differentially edited in females relative to hermaphrodites. This region is not located near any other transcribed elements and lacks an open-reading frame (ORF) of even moderate size. To our knowledge, this transcript would represent the first non-coding mt RNA associated with CMS in plants and is, therefore, an important target for future functional validation studies.
Keywords: Cytoplasmic male sterility; Silene vulgaris; editing; mitochondrion; non-coding RNA; splicing; transcriptome..
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



References
-
- Arrieta-Montiel MP, Mackenzie SA. 2011. Plant mitochondrial genomes and recombination. In: Kempken F, ed. Plant mitochondria. New York:Springer, 65–82.
-
- Bernasconi G, Antonovics J, Biere A, et al. 2009. Silene as a model system in ecology and evolution. Heredity 103, 5–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

