Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
- PMID: 28369529
- PMCID: PMC5907831
- DOI: 10.1093/jmcb/mjx010
Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
Abstract
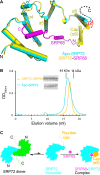
The co-translational targeting or insertion of secretory and membrane proteins into the endoplasmic reticulum (ER) is a key biological process mediated by the signal recognition particle (SRP). In eukaryotes, the SRP68-SRP72 (SRP68/72) heterodimer plays an essential role in protein translocation. However, structural information on the two largest SRP proteins, SRP68 and SRP72, is limited, especially regarding their interaction. Herein, we report the first crystal structures of human apo-SRP72 and the SRP68/72 complex at 2.91Å and 1.7Å resolution, respectively. The SRP68-binding domain of SRP72 contains four atypical tetratricopeptide repeats (TPR) and a flexible C-terminal cap. Apo-SRP72 exists mainly as dimers in solution. To bind to SRP68, the SRP72 homodimer disassociates, and the indispensable C-terminal cap undergoes a pronounced conformational change to assist formation of the SRP68/72 heterodimer. A 23-residue polypeptide of SRP68 is sufficient for tight binding to SRP72 through its unusually hydrophobic and extended surface. Structural, biophysical, and mutagenesis analyses revealed that cancer-associated mutations disrupt the SRP68-SRP72 interaction and their co-localization with ER in mammalian cells. The results highlight the essential role of the SRP68-SRP72 interaction in SRP-mediated protein translocation and provide a structural basis for disease diagnosis, pathophysiology, and drug design.
Keywords: SRP68; SRP72; cancer; crystal structures; protein translocation; protein–protein interaction; signal recognition particle.
© The Author (2017). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, IBCB, SIBS, CAS.
Figures





References
-
- Cingolani G., Petosa C., Weis K., et al. . (1999). Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

