New inhibitor targeting human transcription factor HSF1: effects on the heat shock response and tumor cell survival
- PMID: 28369544
- PMCID: PMC5449623
- DOI: 10.1093/nar/gkx194
New inhibitor targeting human transcription factor HSF1: effects on the heat shock response and tumor cell survival
Abstract
Comparative modeling of the DNA-binding domain of human HSF1 facilitated the prediction of possible binding pockets for small molecules and definition of corresponding pharmacophores. In silico screening of a large library of lead-like compounds identified a set of compounds that satisfied the pharmacophoric criteria, a selection of which compounds was purchased to populate a biased sublibrary. A discriminating cell-based screening assay identified compound 001, which was subjected to systematic analysis of structure-activity relationships, resulting in the development of compound 115 (IHSF115). IHSF115 bound to an isolated HSF1 DNA-binding domain fragment. The compound did not affect heat-induced oligomerization, nuclear localization and specific DNA binding but inhibited the transcriptional activity of human HSF1, interfering with the assembly of ATF1-containing transcription complexes. IHSF115 was employed to probe the human heat shock response at the transcriptome level. In contrast to earlier studies of differential regulation in HSF1-naïve and -depleted cells, our results suggest that a large majority of heat-induced genes is positively regulated by HSF1. That IHSF115 effectively countermanded repression in a significant fraction of heat-repressed genes suggests that repression of these genes is mediated by transcriptionally active HSF1. IHSF115 is cytotoxic for a variety of human cancer cell lines, multiple myeloma lines consistently exhibiting high sensitivity.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
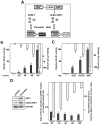

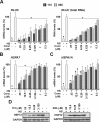

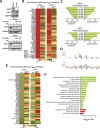

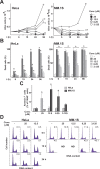
References
-
- Richter K., Haslbeck M., Buchner J.. The heat shock response: life on the verge of death. Mol. Cell. 2010; 40:253–266. - PubMed
-
- Hartl F.U., Bracher A., Hayer-Hartl M.. Molecular chaperones in protein folding and proteostasis. Nature. 2011; 475:324–332. - PubMed
-
- Ananthan J., Goldberg A.L., Voellmy R.. Abnormal proteins serve as eukaryotic stress signals and trigger the activation of heat shock genes. Science. 1986; 232:522–524. - PubMed
-
- Hightower L.E. Cultured animal cells exposed to amino acid analogues or puromycin rapidly synthesize several polypeptides. J. Cell. Physiol. 1980; 102:407–427. - PubMed
-
- McMillan D.R., Xiao X., Shao L., Graves K., Benjamin I.J.. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 1998; 273:7523–7528. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

