Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A
- PMID: 28369571
- PMCID: PMC5435913
- DOI: 10.1093/nar/gkx186
Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A
Abstract
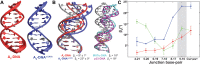
In the canonical DNA double helix, Watson-Crick (WC) base pairs (bps) exist in dynamic equilibrium with sparsely populated (∼0.02-0.4%) and short-lived (lifetimes ∼0.2-2.5 ms) Hoogsteen (HG) bps. To gain insights into transient HG bps, we used solution-state nuclear magnetic resonance spectroscopy, including measurements of residual dipolar couplings and molecular dynamics simulations, to examine how a single HG bp trapped using the N1-methylated adenine (m1A) lesion affects the structural and dynamic properties of two duplexes. The solution structure and dynamic ensembles of the duplexes reveals that in both cases, m1A forms a m1A•T HG bp, which is accompanied by local and global structural and dynamic perturbations in the double helix. These include a bias toward the BI backbone conformation; sugar repuckering, major-groove directed kinking (∼9°); and local melting of neighboring WC bps. These results provide atomic insights into WC/HG breathing dynamics in unmodified DNA duplexes as well as identify structural and dynamic signatures that could play roles in m1A recognition and repair.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Watson J.D., Crick F.H.C.. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. - PubMed
-
- Hoogsteen K. The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1963; 16:907–916.
-
- Abrescia N.G.A., González C., Gouyette C., Subirana J.A.. X-ray and NMR studies of the DNA oligomer d(ATATAT): Hoogsteen base pairing in duplex DNA. Biochemistry. 2004; 43:4092–4100. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

