Robust microbial cell segmentation by optical-phase thresholding with minimal processing requirements
- PMID: 28371011
- PMCID: PMC6585648
- DOI: 10.1002/cyto.a.23099
Robust microbial cell segmentation by optical-phase thresholding with minimal processing requirements
Abstract
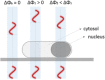
High-throughput imaging with single-cell resolution has enabled remarkable discoveries in cell physiology and Systems Biology investigations. A common, and often the most challenging step in all such imaging implementations, is the ability to segment multiple images to regions that correspond to individual cells. Here, a robust segmentation strategy for microbial cells using Quantitative Phase Imaging is reported. The proposed method enables a greater than 99% yeast cell segmentation success rate, without any computationally-intensive, post-acquisition processing. We also detail how the method can be expanded to bacterial cell segmentation with 98% success rates with substantially reduced processing requirements in comparison to existing methods. We attribute this improved performance to the remarkably uniform background, elimination of cell-to-cell and intracellular optical artifacts, and enhanced signal-to-background ratio-all innate properties of imaging in the optical-phase domain. © 2017 International Society for Advancement of Cytometry.
Keywords: image cytometry; label free; segmentation; single-cell.
© 2017 International Society for Advancement of Cytometry.
Figures

References
-
- Meijering E, Carpenter AE, Peng H, Hamprecht FA, Olivo‐Marin J‐C. Imagining the future of bioimage analysis. Nat Biotech 2016;34:1250–1255. - PubMed
-
- Stephens DJ, Allan VJ. Light microscopy techniques for live cell imaging. Science 2003;300:82–86. - PubMed
-
- Zenobi R. Single‐cell metabolomics: Analytical and biological perspectives. Science 2013;342:1243259. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

