Selective Enzymatic Demethylation of N2 ,N2 -Dimethylguanosine in RNA and Its Application in High-Throughput tRNA Sequencing
- PMID: 28371071
- PMCID: PMC5497677
- DOI: 10.1002/anie.201700537
Selective Enzymatic Demethylation of N2 ,N2 -Dimethylguanosine in RNA and Its Application in High-Throughput tRNA Sequencing
Abstract
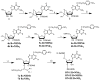
The abundant Watson-Crick face methylations in biological RNAs such as N1 -methyladenosine (m1 A), N1 -methylguanosine (m1 G), N3 -methylcytosine (m3 C), and N2 ,N2 -dimethylguanosine (m22 G) cause significant obstacles for high-throughput RNA sequencing by impairing cDNA synthesis. One strategy to overcome this obstacle is to remove the methyl group on these modified bases prior to cDNA synthesis using enzymes. The wild-type E. coli AlkB and its D135S mutant can remove most of m1 A, m1 G, m3 C modifications in transfer RNA (tRNA), but they work poorly on m22 G. Here we report the design and evaluation of a series of AlkB mutants against m22 G-containing model RNA substrates that we synthesize using an improved synthetic method. We show that the AlkB D135S/L118V mutant efficiently and selectively converts m22 G modification to N2 -methylguanosine (m2 G). We also show that this new enzyme improves the efficiency of tRNA sequencing.
Keywords: AlkB mutant; demethylase; methylguanosine; reverse transcriptase reaction; tRNA sequencing.
© 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
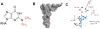


References
-
- Zhu C, Yi C. Angew Chem Int Ed Engl. 2014;53:3659–3662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

