High-Level Resistance of Staphylococcus aureus to β-Lactam Antibiotics Mediated by Penicillin-Binding Protein 4 (PBP4)
- PMID: 28373193
- PMCID: PMC5444179
- DOI: 10.1128/AAC.02727-16
High-Level Resistance of Staphylococcus aureus to β-Lactam Antibiotics Mediated by Penicillin-Binding Protein 4 (PBP4)
Abstract
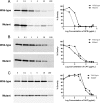
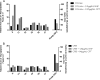
Penicillin-binding protein 4 (PBP4), a nonessential, low-molecular-weight penicillin-binding protein of Staphylococcus aureus, has been implicated in low-level resistance to β-lactam antibiotics, although the mechanism is unknown. Mutations in PBP4 and its promoter were identified in a laboratory-generated mutant strain, CRB, which expresses high-level resistance to β-lactams, including resistance to the new-generation cephalosporins active against methicillin-resistant strains of S. aureus These mutations did not appreciably alter the β-lactam antibiotic binding affinity of purified recombinant mutant PBP4 compared to that of wild-type PBP4. Compared to the susceptible parent strain, COLnex, the CRB strain produces a highly cross-linked cell wall peptidoglycan, indicative of increased transpeptidase activity. The pbp4 promoter mutation of CRB was associated with greatly increased amounts of PBP4 in membranes compared to those in the COLnex parent. Replacement of the native promoter of COLnex with the mutant promoter of CRB resulted in increased amounts of PBP4 in membranes and a highly cross-linked cell wall. PBP4 can be repurposed to provide essential transpeptidase activity in vivo and confer high-level resistance to β-lactam antibiotics, such as ceftobiprole and ceftaroline.
Keywords: PBP4; Staphylococcus aureus; ceftaroline; ceftobiprole; drug resistance mechanisms; mechanisms of resistance; penicillin-binding protein 4; penicillin-binding proteins; β-lactam antibiotics; β-lactams.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, Holden MT, Coleman DC, Goering R, Giffard PM, Skov RL, Zhang K, Westh H, O'Brien F, Tenover FC, Oliveira DC, Boyle-Vavra S, Laurent F, Kearns AM, Kreiswirth B, Ko KS, Grundmann H, Sollid JE, John JF Jr, Daum R, Soderquist B, Buist G. 2012. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother 56:4997–4999. doi:10.1128/AAC.01199-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

